* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Symmetry
Protein design wikipedia , lookup
Structural alignment wikipedia , lookup
Homology modeling wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Cooperative binding wikipedia , lookup
Protein domain wikipedia , lookup
Protein folding wikipedia , lookup
Alpha helix wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein purification wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein structure prediction wikipedia , lookup
Western blot wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
List of types of proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Alcohol dehydrogenase wikipedia , lookup
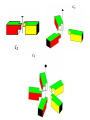
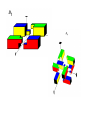
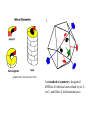
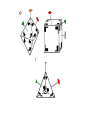
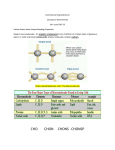
Symmetry is the concept of repetitive arrangements of similar objects in space. In three dimensions, objects may be arranged in a large number of ways - many of these are exhibited in crystal forms. Each possible arrangement is achieved by a combination of simple, basic operations. These include translation, rotation, screw-rotation, mirror, inversion, inversion-rotation, and glide-reflection. The total number of possible ways of arranging copies of the same object to form a repeating lattice in 3D-space is 230. These are the `Space Groups' familiar to crystallographers. Proteins are chiral objects, and cannot be mirror-inverted while remaining the same. Their mirror reflection is different. Thus, many of these arrangements are actually precluded. In fact, proteins may only adopt 65 of the 230 possible 3D space groups. Many of these are observed when we crystallize proteins. In the case of naturally occurring multimers of proteins, other constraints occur which limit the possible arrangements. We are speaking here of individual assemblies of monomer units, creating (usually soluble) complexes which exhibit internal symmetry. Thus, the monomers must associate with van der Waals contact interfaces between the sub-units of the assembly. That is, the sub-units can touch but not intersect. They may interpenetrate only in so far as there exist corresponding `holes' into which `knobs' can be fit. Furthermore, axes of rotational symmetry may not actually pass `through' a protein monomer An icosahedral symmetry, designated I/532 has 60 identical units related by six 5-, ten 3- and fifteen 2-fold rotational axes. Horse liver alcohol dehydrogenase exists as a homodimer or heterodimerthere are two different types of monomer, denoted E and S. However these two forms are near-identical (both consisting of 374 amino acids), as there are only six residues which are different. None of these six occur in the interface region between the two. There are three possible combinations of the subunits: EE, SS and ES; these are therefore isozymes (or isoenzymes). When isolated from liver, 40-60% of the enzyme is of the EE form. Each of the two subunits of the horse enzyme has one binding site for NAD+ and two binding sites for Zn2+. Only one of the zinc ions is involved directly in catalysis. It is ligated by the side chains of Cys 46, His 67, Cys 174 and a water molecule which hydrogen bonds to Ser 48. (If you wish to view this water molecule, download the 8adh PDB structure, which is a monomer without NAD+, as the 6adh dimer includes no water molecules.) The zinc ion is situated at the junction of two binding sites: one of these is a pocket which binds NAD+, while the other is a cleft which binds the substrate (which is one of a variety of aromatic and aliphatic alcohols). Horse liver alcohol dehydrogenase TRAP Trp attenuation protein of Bacillus subtilis The biological unit of this protein is an undecamer (11mer), exhibiting C11 symmetry; however in this particular crystal (1wap) the rings pack back-toback, and there are two rings in the asymmetric unit. Therefore the appearance is of D11 symmetry.. Diagram courtesy of The Protein Structure Group, Chemistry Department, University of York. serum amyloid P-component