* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Affinity Chromatography
Homology modeling wikipedia , lookup
Structural alignment wikipedia , lookup
Immunoprecipitation wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein folding wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein domain wikipedia , lookup
Protein moonlighting wikipedia , lookup
List of types of proteins wikipedia , lookup
Circular dichroism wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Western blot wikipedia , lookup
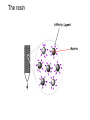
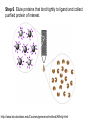
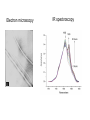
Affinity Chromatography Yongting Wang Jan07 What is AC? • Affinity chromatography (AC) is a technique enabling purification of a biomolecule with respect to biological function or individual chemical structure. • AC is designed to purify a particular molecule from a mixed sample. The resin Affinity Ligand Matrix Examples of tags and ligands • • • • • • His-tag FLAGTM peptide Strep-tag GST tag Maltose binding protein fusion Calmodulin binding protein fusion There are situations where you don’t need a tag. • • • • • • Transition metal ion Monoclonal antibody Biotin Glutathione Amylose Ca2+ Step 1. Loading affinity column. Step 2. Proteins sieve through matrix of affinity beads. Step 3. Proteins interact with affinity ligand with some binding loosely and others tightly. Step 4. Wash off proteins that do not bind. Step 5. Wash off proteins that bind loosely. Step 6. Elute proteins that bind tightly to ligand and collect purified protein of interest. http://www.bio.davidson.edu/Courses/genomics/method/Affinity.html Affinity chromatography applied to recombinant proteins Purity test SDS-PAGE Mass spectrometry N-terminal sequencing, etc. Downstream of protein purification • • • • • Biophysical characterization Biochemical analysis of activities Physiological relevance Pathological mechanisms etc. Example: Human lens crystallins Human gene cloned into pET vector and expressed in E. coli. Diagrams of crystal structure of HgD Trp68 Fluorescence spectroscopy Unfolded Trp156 Native Trp42 N-terminal domain Trp130 C-terminal domain Protein folding mechanisms Electron microscopy IR spectroscopy • In summary, Affinity chromatography is a method that will allow you to purify a particular molecule from a mixed sample so that further investigation of this molecule could be carried out. • Thank you for your attention. Three groups of properties of the target molecule are used in affinity chromatography: 1. Specific binding properties based on biological activity like: - Enzyme active sites - Receptor binding sites - Antibody binding sites etc. These are used together with the natural ligand or an analogue of it. Sometimes the analogue has a broader specificity and can be used for group separations. 2. Naturally occurring prosthetic groups like: polysaccharides etc. Such properties normally allow group separations only. 3. Molecules equipped with an affinity tag like: - Glutathione-S-Transferase (GST) - Oligo histidine etc. This group of properties is used almost exclusively for recombinant fusion proteins.