* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Peptide Bonds - Newcastle University
Bimolecular fluorescence complementation wikipedia , lookup
Protein design wikipedia , lookup
Protein purification wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein domain wikipedia , lookup
Circular dichroism wikipedia , lookup
Homology modeling wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Western blot wikipedia , lookup
Protein folding wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
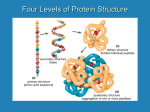
Amino acids join together via condensation reactions to form peptide bonds. Chains of amino acids are called polypeptides. The order of amino acids in these chains is specific and this determines the primary structure of a protein. Peptide Bonds Peptide bond formation is a nucleophilic addition-elimination reaction. The nitrogen lone pair of one amino acid attacks the carbonyl carbon of another, leading to the formation of a peptide bond. This is the addition step; the elimination stage of the reaction is shown below. Follow the curly arrows and the movement of hydrogen atoms until you reach the dipeptide and eliminated water molecule. Remember that the peptide bond is rigid and planar because it is stabilised by delocalisation of the nitrogen lone pair with the carbonyl oxygen. Primary structure Two amino acids joined together are called a dipeptide. The condensation reaction you have just seen can repeat so any more amino acids can add together in the same way, forming a long chain called a polypeptide. For each protein, the order of amino acid residues is specific and different – it is this order that gives the protein its primary structure. Remember that the order of amino acid residues determines how the protein will eventually fold – the shape it will take. This is important to the protein’s function – how it works. One single change to the amino acid sequence can completely change the proteins ability to function – that’s how important it is! Imagine these amino acids join together like you have seen on the previous page: Leucine – cysteine – methionine – valine – glycine The primary structure of this protein could be represented as: LEU – CYS – MET – VAL – GLY At the left hand side of the protein chain, there will be an amino acid with an unchanged –NH2 group. In this example it is leucine, and this is called the protein’s N-terminal. At the other end of the protein chain is and unchanged –COOH group, in this example it belongs to glycine. This is known as the Cterminal. Or like this: LCMVG To save time, each amino acid has a three-letter and one-letter code. C=O C-terminal Nitrogen N-terminal The 3D representation of LCMVG – notice the repeating amino acid units and the R-group side chains extending from either side of the polypeptide chain. Produced by Lucy Jakubecz at Newcastle University as part of an MChem project.