* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Globular Protein Structure
Silencer (genetics) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Signal transduction wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Expression vector wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Gene expression wikipedia , lookup
Point mutation wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Protein purification wikipedia , lookup
Biochemistry wikipedia , lookup
Western blot wikipedia , lookup
Metalloprotein wikipedia , lookup
Interactome wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Homology modeling wikipedia , lookup
Proteolysis wikipedia , lookup
Protein–protein interaction wikipedia , lookup
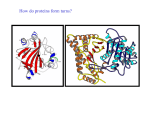
Karolinska Institutet & Södertörns högskola Structural biochemistry Computer lab 3 HT05 Globular Protein Structure – Structural Organization & Motifs DNA-Protein/Drug Interactions Introduction The basic functional unit of protein tertiary structure is the domain. In its basic form, a domain is a structurally or functionally distinct parts of the protein. Small globular proteins (e.g., ribonuclease A) usually have only one domain whereas larger proteins (remember proteins can have molecular weights of > 1 x 106) can have as many as several dozen. The term domain is very often used (carelessly) when speaking about protein structure. Therefore it is important to understand the different uses of this fundamental unit of protein organization. Basically all definitions can be divided into two classes: structural domains and functional domains. Structural Domain: Independent compactly folded (globular) structures. Structural domains should have their own hydrophobic core and can be made up of one or several polypeptide chains. One must be careful not to assume that structural domains are in themselves capable of existing outside the context of the entire protein. It is possible to define folding domains ONLY through experiment (e.g., proteinase digestion or cloning). Just because it looks autonomous (independent), it doesn’t mean that it is. Functional Domain: These are regions of structure associated with a particular function. They may correspond to one or several structural domains. Very often the functional domain is at an interface between two structural domains. DNA-Protein/Drug Interactions: Unfortunately, there is no general rule that governs DNAbinding by proteins and small molecules. Nature has apparently found multiple solution to this very important problem. However as usual, there are underlying themes which you will notice that relates stability, function, and specificity Objectives • • • To see some examples of how proteins can be organized into domains as a prelude to their further dissection into structural units which permits a classification of protein structure. To identify some basic, recurring structural motifs found in proteins. To identify some basic structural features of nucleic acid-protein/drug interactions. RasMol Scripts In this lab you will use RasMol scripts, which have already been prepared, to help with the display of certain features. The scripts are regular text files and can be downloaded from http://www.biosci.ki.se/events/courses/sbb/ht05.html -1- Karolinska Institutet & Södertörns högskola Structural biochemistry Computer lab 3 HT05 Part 1 Amphiphilic Structures Quite a few structures conform to the low-resolution view of protein structure as an oil drop or micelle. Proteins can be can be considered as amphiphilic (containing both hydrophilic and hydrophobic groups which through folding segregate into larger polar and apolar regions) on many different structural levels. We will look at the following proteins Myoglobin (1vxa), Green fluorescent protein (1gfl), Porin (2por): Exercises: 1. Use your browser and download these coordinate files. Try to identify the amphiphilic organization in these structures. Hint: RasMol allows you to “select hydrophobic” or “select not hydrophobic” 2. In general do these proteins conform to the hydrophobic in and hydrophilic out principle? 3. Why does porin seem to violate the oil drop model? 4. With pencil and paper, draw helical wheel diagrams for those helices that you identify. Also draw a schematic for the amphiphilic beta structures. The link http://www.site.uottawa.ca/~turcotte/resources/HelixWheel/ may help with the helical wheels. Remember the amino acid sequences can be obtained in oneletter format through a link obtained on the PDB data page for that coordinate file. Some examples of domains Visit the PDB web page (linked from the course homepage) and obtain the following coordinate files. 2adm 1pky 1rpl Exercise: 1. Use RasMol to create a display of the protein that will allow you to identify visually structural domains. An example of this is the image on page 1 of this lab. Questions: 1. Does the coordinate file contain one or several polypeptide chains? The RasMol menu option colors “chain” can help here. 2. Do the chains have the same sequence? That is to say are there several structures with the same sequence in the file? 3. How many structural domains exist in each. Are the domains similar/identical or different? Visit to the CATH protein classification home page One useful classification of all known structures is the CATH database. We will be using the CATH classification of proteins in the coming lectures/labs. Take this opportunity to browse the homepage and obtain an overview of the organization of this important database. http://www.biochem.ucl.ac.uk/bsm/cath/ -2- Karolinska Institutet & Södertörns högskola Structural biochemistry Computer lab 3 HT05 Exercise: 1. Find the classification of the protein you have been looking at in detail, ribonuclease A (5rsa). 2. Notice as you browse that classification is based on structural organization. Part 2 Structural Motifs in Globular Proteins The files that you need in parts 2 and 3 of the lab are in a self-extracting archive, which you find from the course webpage. Helix bundle motif Helix bundles are a common structural motif (also called architecture) that has been extensively document. They can be observed both independently and as components of larger folding units. The basic motif comprises four helices packed together in a variety of different ways, forming a hydrophobic core. In the most common bundles, the helices that are adjacent in the amino acid sequence are also adjacent in the three dimensional structure, forming the so-called up-down-up-down four helix bundle. The side chains of each helix in the bundle are arranged such that the hydrophobic side chains are buried between the helices and the hydrophilic side chains are exposed on the surface of the bundle. Examples of two different types of bundles are the proteins E. coli cytochrome B256 and human hemoglobin. Note that the RasMol command to restrict to a particular chain is: RasMol> restrict *B Exercises: 4. An example of an anti-parallel 4 helix bundle is E. coli cytochrome B256 (256b.pdb, script 256b_1.txt). The protein contains an iron-containing heme prosthetic group (yellow and grey). What is the angle between the helices? Are the helices amphiphilic? What advantages does the antiparallel arrangement present? 5. Human hemoglobin (4hhb.pdb) contains four protein subunits, each with a similar tertiary structure. The structure is similar to the myoglobin you looked at previously. Each subunit also contains a heme group. Before running the script, look at the quaternary structure of hemoglobin. The script 4hhb_2.txt selects the "B" subunit and colors 4 helices red, the heme yellow and iron grey. Parallel β sheet motifs Parallel β strands are most often arranged in closed barrels or flat sheets. In each case, the parallel strands are associated with α helices. The fundamental unit common to both arrangements is the β-α-β motif. This motif comprises a β strand-loop- α helix-loop-strand arrangement, with the strands arranged parallel to the connecting helix. The α helix packs against the beta strands and thus buries the hydrophobic side chains in these strands. The joining loops regions in the motif can vary in length from one residue to more than 100 residues. Interestingly, essentially all of these β-α-β motifs have the same handedness, they are right handed (inset figure). -3- Karolinska Institutet & Södertörns högskola Structural biochemistry Computer lab 3 HT05 A Rossmann Fold is a β-α-β-α-β motif in which the strands form a parallel β sheet with (+1x, +1x) connectivity, and the helices lie anti-parallel to the strands on one side of the sheet. Two Rossman folds, wound in opposite directions create a dinucleotide binding unit found in many enzymes, specifically in nucleotide binding proteins (hence its alternative name mononucleotide-binding motif). Exercises: 1. The classical example of a β-α-β motif which forms a barrel is triosephosphate isomerase (1tim.pdb; note the coordinate file contains two molecules. We will look at only chain A). Identify and count the β-α-β motifs. This topology is referred to as a singularly wound structure. Draw a topology diagram of the structure. 2. Not all parallel sheets form barrels. There is a large family of proteins which have an open twisted parallel sheet formed from 5 strands. An example of this motif is the protein flavodoxin (2fx2.pdb). The central parallel sheet is “sandwiched” between helices. This type of structure is called a 3–layer sandwich. Identify the three layers and the β-α-β motifs. The script 2fx2_1.txt color codes the first motif. This structure is referred to as doubly wound. See if you can identify the flavin binding site. 3. There are three β-α-β motifs in the bacterial proteinase subtilisin (1cse.pdb). The script 1cse_1.txt colors one of the motifs, and the script 1cse_2.txt colors the active site magenta .What is unusual about this β-α-β motif? Active site sidechains are colored magenta. Why is this exception present? Antiparallel β sheet motifs Antiparallel β sheet containing proteins are the most functionally diverse family of structures. Antiparallel β structures can be as simple as a simple hairpin structure to very complicated superbarrels. Of the many different topologies possible, three are occurring very frequently. These are the up-and-down β barrels, Greek Keys, and jelly roll motifs. The up-and-down structures are the simplest to visualize. The topologies are shown in the figure. They can exist as flat sheets or barrels. The Greek Key is a four stranded beta sheet motif which is characterized by +3, -1, -1 connectivities in a 2-dimensional schematic diagram of a protein structure (see inset topological diagram). In 3-dimensions the motifs adopts a variety of structures. This motif can be considered as a chain folded in half, then twisted around to form a pattern like that commonly seen on Greek vases. Greek keys can occur with all four participating strands in the same beta sheet or in two different sheets. A four-stranded Greek key motif can be defined by the following rules: four sequential strands must be either in one or at most two different β sheets within any one sheet the participating strands must be hydrogen bonded together hydrogen-bonded strands within one sheet must be antiparallel strands 1 and 4 must be hydrogen bonded . The Greek Key motif has only one connection across the barrel. The Jelly Roll barrel is a fold topology that classically consists of four Greek key motifs that adopt an eightstranded beta sandwich structure. In this fold the hydrogen -4- Karolinska Institutet & Södertörns högskola Structural biochemistry Computer lab 3 HT05 bonding pattern between adjacent strands is broken in two places and as a consequence the structure comprises two four stranded beta sheets. Both sheets are purely antiparallel, with strands adjacent in sequence appearing in different sheets with the exception of the fourth and fifth strands which are in the same sheet. This leads to a structure with only one haipin, all other beta-beta connections being arches. There are four connections that cross the barrel, two on top and two on the bottom. When the structure is depicted in two-dimensions the origin of its name is apparent; the structure resembles a jelly roll. In theory there could be four types of jellyroll folds, two have a right-handed superhelical twist and two have a left handed twist: but as with the β-α-β motif, only right-handed Greek key, and jellyroll structures are actually observed in globular proteins. In practice the jelly roll may be varied by the addition or deletion of secondary structure elements. Exercises: 1. Bovine pancreatic trypsin inhibitor (5pti.pdb) is an example of a very simple β hairpin. You have seen this protein last time. Take another look and identify the β hairpin structure. A β hairpin connection is one in which the polypeptide chain reenters the same end of the beta sheet it left. The strands lie adjacent in the sheet so that a β hairpin always has a +1 or -1 nomenclature, with the strands hydrogen bonded together. 2. Retinol-binding protein (1cbs.pdb) is an example of a protein containing an upand-down β barrel. The barrel consists of some 10 strands which can be considered as two 5-stranded sheets. Identify the two halves of the barrel. Make a topology diagram of the structure. Retinoic acid binds inside the barrel. The script 1cbs_1.txt may help. 3. Neuraminidase is viral protein of 4 identical chains of about 470 amino acids. Its function is to cleave sialic acid groups from the carbohydrates attached to membrane proteins. Selective proteinase treatment cleaves off a portion called the headpiece (residues 84-469). The structure of a single headpiece is given in the coordinate file (2bat.pdb). One motif is color coded with the script 2bat_1.txt. Note the sialic acid substrate is colored red). What topology does it have? How many repeating motifs make up the headpiece? Part 3 DNA-Protein/Drug Interactions DNA-Protein Interactions In the follow exercises you will get first-hand experience with two different modes of binding DNA used by proteins. Trp Repressor/Operator Complex The E. coli Trp repressor is a dimer of intertwined monomers which controls the operon for synthesis of L-tryptophan. It is a good example of a negative feedback system. When tryptophan is abundant, the repressor binds tryptophan which causes a conformational change enabling DNA binding and shutdown of transcription. Surprisingly, despite the sequence specific DNA recognition, the repressor makes no direct interactions with the DNA bases. It uses water-mediated recognition of bases in the major groove. It distorts the DNA making 24 direct and 6 water-mediated contacts to the phosphate backbone. In this complex, it appears that the flexibility of the DNA is essential for recognition. -5- Karolinska Institutet & Södertörns högskola Structural biochemistry Computer lab 3 HT05 Exercises: 1. The RasMol script 1tro_1.txt will load the coordinate file and demonstrate some essential interactions. Note: there are several “pauses” to allow you to look around. To move on, press the spacebar. The command window describes the view. 2. Identify the helix-turn-helix motif. 3. Measure the distance between the two recognition helices. Zinc finger By far the most common eukaryotic gene regulatory proteins contain zinc fingers. There are basically three different versions of the finger motif, yet all insert an a helix into the major groove. Very often the zinc fingers are located in tandem arrangements. This exercise will focus on a complex between the mouse zinc finger Zif268 and DNA. The biological function of Zif268 is not yet known, but the protein is suspected to be an activator of transcription, because it is made in response to certain growth factors which induce the cell to divide. Exercises: 4. Look at the structure of the mouse Zif268 zinc finger. The RasMol script 1aay_1.txt will load the coordinate file and get you started. 5. Notice the role of the highlighted Arg, His and Asp residues. 6. Are there any water-mediated contacts in this complex? 7. How is the α-helix held against the β-strand in each finger? 8. Identify the three domains in the amino acid sequence of the zinc finger. Highlight the residues involved in zinc binding, specific and non-specific DNA binding. 1 MERPYACPVE SCDRRFSRSD ELTRHIRIHT GQKPFQCRIC MRNFSRSDHL 51 TTHIRTHTGE KPFACDICGR KFARSDERKR HTKIHLRQKD -6- Karolinska Institutet & Södertörns högskola Structural biochemistry Computer lab 3 HT05 DNA-Drug Interactions Most DNA-binding drugs act by either binding to the minor groove or intercalating. In this example one successful drug will be examined. Hoechst 33258 Hoechst 33258 (minor groove binder) is a cationic antibiotic and chromosome stain which binds to the minor groove of adenine-thymine rich DNA segments. Hoechst 33258, excites in the near UV (350 nm) and emits in the blue region (450 nm) and has a fluorescent sensitivity of approximately 5 ng/mL. Exercises: 1. Look at Hoechst 33258 with the script 1d45_1.txt. Notice the specific H-bond interactions with the bases A6 and T7. The drug/DNA complex is stabilized by single or bifurcated hydrogen bonds between the two N-H hydrogen-bond donors in the benzimidazole rings of Hoechst and adenine N3 and thymine O2 acceptors in the minor groove. 2. Notice how O4 of the DNA deoxyribose ring interacts with the π electrons of the drug. 3. A general preference for AT regions is conferred by electrostatic potential and by narrowing of the walls of the groove. 4. Does solvent appear to play a role in this complex? -7- Karolinska Institutet & Södertörns högskola Structural biochemistry Computer lab 3 HT05 Answer sheet Amphiphilic structures In general do these proteins conform to the hydrophobic in and hydrophilic out principle? Why does porin seem to violate the oil drop model? Domain examples: 2adm 1pky 1rpl Does the coordinate file contain one or several polypeptide chains? Do the chains have the same sequence? That is to say are there several structures with the same sequence in the file? How many structural domains exist in each. Are the domains similar/identical or different? CATH Classification of 5RSA? Helix bundle motif What is the angle between the helices? Are the helices amphiphilic? What advantages does the antiparallel arrangement present? Parallel beta sheet What is unusual about this β-α-β motif? Why is this exception present? Antiparallel beta sheet What is the topology of 2bat? How many motifs are there in the headpiece? DNA/Protein Measure the distance between the two recognition helices. Zinc finger Are there any water-mediated contacts in this complex? How is the α-helix held against the β-strand in each finger? Highlight the residues involved in zinc binding, specific and non-specific DNA binding. Hoechst Does solvent appear to play a role? -8- MERPYACPVE SCDRRFSRSD ELTRHIRIHT GQKPFQCRIC MRNFSRSDHL TTHIRTHTGE KPFACDICGR KFARSDERKR HTKIHLRQKD