* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Abstract - Plant Sulfur Network
Gene desert wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gene expression wikipedia , lookup
Proteolysis wikipedia , lookup
Biosynthesis wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Biochemical cascade wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene regulatory network wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Ridge (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Molecular evolution wikipedia , lookup
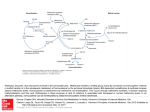
SULFUR METABOLISM IN ASPERGILLUS NIDULANS: PATHWAYS AND REGULATION1 Andrzej Paszewski, Renata Natorff, Marzena Sieńko, Jerzy Brzywczy and Irmina Lewandowska Department of Genetics, Institute of Biochemistry and Biophysics PAS, Pawińskiego 5a, Warszawa, Poland Abstract -Figures 2, 3 and 4 are not cited in the text – please cite them in the correct places -comments to Fig1 and its legend: (1) what is in italics – presumably genes names but explain that there are genes names in the legend, (2)“cB” should be probably “cysB” etc., (3) show where is SAH lyase if possible –this would help to explain Fig. 4., (4) replace “—“ with “2-“ -comments to Fig.4 and its legend: perhaps after explaining in Fig.1 where is SAH lyase it would be possible to have the legend as corrected. The final product of the sulfate assimilation pathway, sulfide, is incorporated into serine or homoserine carbon chain, forming cysteine or homocysteine, respectively. There are three types of pathway organization by which sulfur amino acids are synthesized and interconverted. Plants, Schizosaccharomyces pombe and Enterobacteriaceae synthesize de novo cysteine, which is sequentially converted to methionine. Saccharomyces cerevisiae makes homocysteine, which serves as a precursor of both cysteine and methionine. Only in Aspergillus nidulans there are alternative pathways for cysteine and homocysteine de novo synthesis. Impairment of both pathways results in cysteine auxotrophy. It was found, however, that the route involving homocysteine synthase, cystathionine β-synthase and cystathionine γ-lyase (Fig. 1, steps 11, 12, 13) functions efficiently when the main pathway of cysteine synthesis from O-acetylserine is impaired. In consequence, mutations in the cysA and cysB genes lead to elevated synthesis of homocysteine by homocysteine synthase so they suppress mutations in the metB, metG and metA genes. In fact, mutations in cys genes are readily isolated as suppressors of mutations in the met genes. Another types of isolated suppressor mutations, exerting the same effect as mutations in cys genes, are mutations in regulatory genes. Some of them are located in the four negative-acting scon genes (Natorff et al. 1993), others in the positive-acting metR gene encoding a sulfate assimilation transcriptional activator, which belongs to the bZIP family (Natorff et al. 2003). Scon and metR genes constitute the sulfur metabolite repression (SMR) system, which controls the expression of genes encoding the sulfate assimilation pathway and Sulfur Metabolism in Higher Plants, pp. xx-xx Edited by A. Sirko et al. 2009 Backhuys Publishers, Leiden, The Netherlands homocysteine synthase. Two scon genes encode components of a multiprotein ubiquitin ligase of SCF-type (Skp1-Cullin-Fbox). The A. nidulans SconB protein (Natorff et al. 1998) is an orthologue of S. cerevisiae Met30p that determines specificity of SCF ligase. It contains an F-box motif that interacts with the Skp1 adaptor protein encoded by A. nidulans sconC (Piotrowska et al. 2000). The S. cerevisiae orthologue of SconC (Skp1p) is an essential subunit of SCF complexes. SCF ligases are elements of the ubiquitin proteasome system that regulates many cellular processes, including nutrient sensing. Under conditions of cysteine excess, the level or activity of the MetR protein is negatively controlled by the SCFSconB ligase. Mutations in the scon genes lead to loss of sulfur metabolite repression. In this way they cause overproduction of sulfur compounds from sulfate even in the presence of cysteine or methionine, probably as a result of inefficient degradation of the MetR protein. Fig. 1. An outline of sulfur containing amino acid metabolism and folate metabolism in Aspergillus nidulans. Enzymes: 1, sulfate permease; 2 , ATP-sulfurylase; 3, AdoPS-kinase; 4, PadoPS-reductase: 5, sulfite reductase; 6, serine transacetylase; 7, cysteine synthase (Oacetylserine sulfhydrylase); 8, homoserine transacetylase; 9, cystathionine -synthase; 10, cystathionine -lyase; 11, homocysteine synthase; 12, cystathionine -synthase; 13, cystathionine -lyase; 14 , dihydrofolate reductase; 15, serine hydroxymethyltransferase; 16, methylenetetrahydrofolate reductase; 17, methionine synthase; 18, S-adenosyl-methionine synthase; 19, S-adenosylhomocysteine hydrolase. AdoMet, S-adenosylmethionine; AdoHcy, Sadenosylhomocysteine; AdoPS, adenosine 5-phosphosulfate; PadoPS, adenosine 3-phosphate 5phosphosulfate; CH2H4PteGlun, methylenetetrahydrofolylpolyglutamate; CH3H4PteGlun, 5-methyltetrahydrofolylpolyglutamate. Fig. 2. Mutations in the cysB, sconB, sconC and metR genes suppress metB3 and metA17 mutations. Growth of mutants on minimal medium with (or without) 0,25 mM methionine. Fig. 3. Ubiquitination of the MetR protein by SCFSconB ligase at high concentration of cysteine - a low-molecular weight effector of this complex. Ubiquitinated MetR is directed to degradation or inactivated. Losses of function mutations in the metR gene are epistatic to mutations in the negative-acting scon genes and lead to methionine auxotrophy. Dominant-type mutations in the metR gene (MetRd) lead to suppression of mutations in the metA, metB and metG genes. In these mutants a conserved N-terminal phenylalanine (F48) of MetR is changed to serine or leucine. Mutated MetR(F48S) may be more resistant to degradation. Thus, the efficient synthesis of homocysteine (via homocysteine synthase) can be achieved by three different classes of mutations: i) Block in cysteine synthesis (lowered level of regulatory effector), cysA and cysB mutations; ii) SCF complex damage (MetR protein excess), sconB and sconC mutations; iii) MetR protein stabilization – MetRd mutations Both scon and metR ortologues are present in all filamentous fungal species for which genomic sequences are available. Fig. 4. Homocysteine up-regulates genes that encode enzymes involved in homocysteine metabolism: A) metH, metA, metF, mecA, mecB, mecC, and SAH lyase encoding gene. B) folylpolyglutamate synthase (FPGS) and dihydrofolate synthase (DHFS) encoding genes. Northern analysis of the wild type strain grown in minimal medium suplemented with Lhomocysteine (1, 2 or 3 mM), or with L-methionine (5 mM). Homocysteine occupies a branch point in methionine, cysteine, and S-adenosylmethionine metabolism. Elevated levels of homocysteine have various deleterious effects. Cells maintain a low level of homocysteine mainly by means of two metabolic pathways. The first is conversion to cysteine, a precursor of glutathione, the major redox regulating metabolite of the cell. The second is methylation to methionine catalyzed by methionine synthase in a reaction that requires methyltetrahydrofolate. About half of homocysteine formed is conserved by remethylation to methionine in the "methionine cycle". The other half is irreversibly converted to cysteine by cystathionine -synthase and cystathionine -lyase. We have shown that homocysteine up-regulates genes encoding enzymes involved in methionine synthesis: metH coding for methionine synthase (Kacprzak et al. 2003) as well as metA and metF genes encoding methylenetetrahydrofolate reductase isoenzymes which synthesize methyltetrahydrofolate (Sieńko et al. 2007). Homocysteine induces also mecA and mecB genes encoding cystathionine -synthase and cystathionine -lyase necessary for its conversion to cysteine. Recently we have shown that genes encoding methionine catabolic enzymes, S-adenosylmethionine synthase (mecC) and S-adenosylhomocysteine hydrolase, are up-regulated by homocysteine as well as the two genes encoding folate cycle enzymes: folylpolyglutamate synthase and dihydrofolate synthase. Since higher concentrations of homocysteine are toxic to the cell, derepression of these enzymes by homocysteine is an effective means to remove an excess of this amino acid. We call the set of genes up-regulated by homocysteine the „homocysteine regulon”. However, the molecular mechanism of this regulation is not yet elucidated. References Kacprzak, M.M., Lewandowska, I., Matthews, R.G., and Paszewski, A. 2003. Transcriptional regulation of methionine synthase by homocysteine and choline in Aspergillus nidulans. Biochem. J. 376: 517–524. Natorff, R., Balińska, M., and Paszewski, A. 1993. At least four regulatory genes control sulphur metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 238: 185-192 Natorff, R., Piotrowska, M., and Paszewski, A. 1998. The Aspergillus nidulans sulphur regulatory gene sconB encodes a protein with WD40 repeats and an F-box. Mol. Gen. Genet. 257:.255-263 Natorff, R., Sieńko, M., Brzywczy, J., and Paszewski, A. 2003. The Aspergillus nidulans metR gene encodes a bZIP protein which activates transcription of sulphur metabolism genes. Mol. Microbiol. 49: 1081-1094 Piotrowska, M., Natorff, R., and Paszewski, A. 2000. sconC, a gene involved in the regulation of sulphur metabolism in Aspergillus nidulans, belongs to the SKP1 gene family. Mol. Gen. Genet. 264: 276-282 Sieńko, M., Natorff, R., Zieliński, Z., Hejduk, A., and Paszewski, A. 2007. Two Aspergillus nidulans genes encoding methylenetetrahydrofolate reductases are up-regulated by homocysteine. Fungal. Genet. Biol. 44: 691-700.