* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download A-13-LinkageAnalysis
Biology and consumer behaviour wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genetic drift wikipedia , lookup
Fetal origins hypothesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Y chromosome wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Neocentromere wikipedia , lookup
Genome evolution wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Population genetics wikipedia , lookup
Genomic imprinting wikipedia , lookup
X-inactivation wikipedia , lookup
Designer baby wikipedia , lookup
Gene expression programming wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome (book) wikipedia , lookup
Public health genomics wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Microevolution wikipedia , lookup
Gene Hunting: find genes responsible for a given disease
Main idea: If a disease is statistically linked with a marker
on a chromosome, then tentatively infer that a gene causing
the disease is located near that marker.
.
Some slides were prepared by Ma’ayan Fishelson, some by Nir, and most are
mine. I have slightly edited all slides.
Human Genome
Most human cells contain
46 chromosomes:
2 sex chromosomes
(X,Y):
XY – in males.
XX – in females.
22 pairs of
chromosomes named
autosomes.
2
Sexual Reproduction
egg
ביצית
sperm
תא זרע
gametes
zygote
ביצית מופרית
תאי מין
3
Chromosome Logical Structure
Locus – the location of genes
or other markers on the
chromosome.
Allele – one variant form (or
state) of a gene/marker at a
particular locus.
Locus1
Possible Alleles: A1,A2
Locus2
Possible Alleles: B1,B2,B3
4
Genotypes versus Phenotypes
At each locus (except for sex chromosomes) there are 2
genes. These constitute the individual’s genotype at the
locus.
The expression of a genotype is termed a phenotype.
For example, hair color, weight, or the presence or
absence of a disease.
5
Recombination Phenomenon
A recombination between
2 genes occurred if the
haplotype of the individual
contains 2 alleles that
resided in different
haplotypes in the
individual's parent.
(Haplotype – the alleles at
different loci that are received by
an individual from one parent).
6
An example - the ABO locus.
The ABO locus
determines detectable
Phenotype
Genotype
antigens on the surface
A
A/A, A/O
of red blood cells.
The 3 major alleles
B
B/B, B/O
(A,B,O) interact to
AB
A/B
determine the various
ABO blood types.
O
O/O
O is recessive to A and
B. Alleles A and B are
Note that the listed genotypes are unordered
codominant.
(we don’t know which allele is from the father
and which one is from the mother).
7
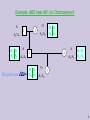
Example: ABO near AK1 on Chromosome 9
O
A
A1/A1
A O
A1 A2
Recombinant
2
1
O O
A2 A2
A2/A2
A
A
A1/A2
4
3
O O
A1 A2
O
A2/A2
A O
A2 | A2
5
A1/A2
8
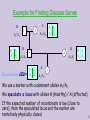
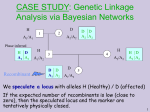
Example for Finding Disease Genes
A
H
A1/A1
H A
A1 A2
Recombinant
2
1
A A
A2 A2
A2/A2
H
H
A1/A2
4
3
A A
A1 A2
A
A2/A2
H |A
A2 | A2
5
A1/A2
We use a marker with codominant alleles A1/A2.
We speculate a locus with alleles H (Healthy) / A (affected)
If the expected number of recombinats is low (close to
zero), then the speculated locus and the marker are
tentatively physically closed.
9
The method just described is called
genetic linkage analysis. It uses
the phenomena of recombination in
families of affected individuals to
locate the vicinity of a disease gene.
10
Comments about the example
Often:
Pedigrees are larger and more complex.
Not every individual is typed.
There are more markers and they have more than
two alleles.
Recombinants cannot always be determined.
11
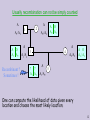
Usually recombination can not be simply counted
A
A
A1/A1
A O
A1 A2
Recombinant ?
Sometimes !
2
1
A O
A 2 A2
A2/A2
A
A
A1/A2
4
3
? ?
A1 A2
A
A2/A2
A O
A2 | A2
5
A1/A2
One can compute the likelihood of data given every
location and choose the most likely location.
12
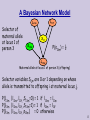
A Bayesian Network Model
L11f
L11m
Selector of
maternal allele
at locus 1 of
person 3
X11
S13m
P(s13m) = ½
L13m
Maternal allele at locus 1 of person 3 (offspring)
Selector variables Sijm are 0 or 1 depending on whose
allele is transmitted to offspring i at maternal locus j.
P(l13m | l11m, l11f,,S13m=0) = 1 if l13m = l11m
P(l13m | l11m, l11f,,S13m=1) = 1 if l13m = l11f
P(l13m | l11m, l11f,,s13m) = 0 otherwise
13
L11m
L12m
L11f
L12f
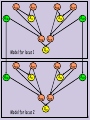
Probabilistic model for two loci
X11
S13m
X12
L13f
L13m
X13
Model for locus 1
L21m
S23m
L22m
L21f
X21
X22
L22f
S23f
L23f
L23m
Model for locus 2
S13f
X23
14
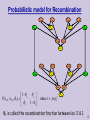
Probabilistic model for Recombination
L11m
L12m
L11f
X11
S13m
L12f
X12
S13f
L13f
L13m
X13
L21m
S23m
L22m
L21f
X21
X22
S23f
L23f
L23m
2
1 2
P( s23t | s13t , 2 )
where t {m,f}
1 2
2
L22f
X23
θ2 is called the recombination fraction between loci 2 & 1.
15
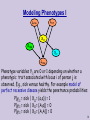
Modeling Phenotypes I
L11f
L11m
X11
S13m
Y11
L13m
Phenotype variables Yij are 0 or 1 depending on whether a
phenotypic trait associated with locus i of person j is
observed. E.g., sick versus healthy. For example model of
perfect recessive disease yields the penetrance probabilities:
P(y11 = sick | X11= (a,a)) = 1
P(y11 = sick | X11= (A,a)) = 0
P(y11 = sick | X11= (A,A)) = 0
16
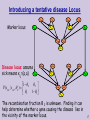
Introducing a tentative disease Locus
L11m
Marker locus
L12m
L11f
X11
S13m
L12f
X12
S13f
L13f
L13m
X13
Disease locus: assume
S
sick means xij=(a,a)
23m
2
1 2
P( s23t | s13t ' , 2 )
1
2
2
L21m
L22m
L21f
X21
L22f
X22
S23f
L23f
L23m
X23
The recombination fraction θ 2 is unknown. Finding it can
help determine whether a gene causing the disease lies in
the vicinity of the marker locus.
17
SUPERLINK
Stage 1: each pedigree is translated into a Bayesian network.
Stage 2: value elimination is performed on each pedigree (i.e.,
some of the impossible values of the variables of the network are
eliminated).
Stage 3: an elimination order for the variables is determined,
according to some heuristic.
Stage 4: the likelihood of the pedigrees given the θ values is
calculated. This is done by by performing variable elimination
according to the elimination order determined in stage 3.
18