* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download SB 2.0 poster
Histone acetylation and deacetylation wikipedia , lookup
Cellular differentiation wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
List of types of proteins wikipedia , lookup
Signal transduction wikipedia , lookup
Hedgehog signaling pathway wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Gene regulatory network wikipedia , lookup
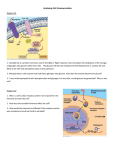
Re-engineering transcriptional control in the yeast pheromone response pathway Alex 1 Mallet & Drew 2 Endy MIT Computational and Systems Biology1 & Biological Engineering2 Abstract Feedback in the pathway The pheromone response pathway in S.cerevisiae is regulated on multiple levels and timescales, and by many biochemical mechanisms. In particular, several proteins that are directly involved in signal transduction through the pathway, as well as several positive and negative regulators of the pathway, are transcriptionally upregulated in response to pheromone. Opening the feedback loops Replacement Promoters STE12-mediated transcriptional feedback Constitutive promoters •Library of TEF promoters4 •Allow varying expression level over 2 orders of magnitude •GPD, TEF, ADH1, BMH2, ACT1, CYC •Capable of producing ~1000 – 90,000 molecules/cell, depending on protein3 The transcription factor STE12 is activated in response to pheromone and increases transcription of several proteins involved in the pathway over their basal expression levels. These proteins, in turn, exert regulatory control on each other, as depicted below. STE12 thus acts as a key component contributing to a set of interlocking positive and negative feedback loops. Previous work has focused on the effects of deleting or overexpressing proteins involved in propagating and modulating the signal. While data from these experiments sheds some light on the impact of varying protein levels, it does not represent a systematic exploration of the importance of transcriptional regulation of pathway genes in response to pheromone. Here, we describe work that explicitly addresses the question whether specific instances of pheromone-dependent transcriptional feedback loops are necessary for proper pathway function. Regulatable promoter •Combination of artificial promoter based on Tetracycline repressor and artificial transcription factor rtTA(S2) 5 •rtTA(S2) binds DNA in the presence of doxycycline, allowing induction of transcription by addition of doxycycline •Allows varying expression level over 3 orders of magnitude, with unimodal cell response and low noise levels6 Determining promoter sequences to replace STE2 Transcriptional upregulation Post-translational downregulation Specifically, we are working to replace the promoters of pheromone-inducible pathway genes, both singly and in combination, with constitutive and exogenously regulatable promoters. These custom promoters will allow us to explore pathway response in the absence of transcriptional feedback, via regulatable expression of some of the major contributors to signal transmission and control. GPA1 SST2 Signal transduction Design rules: • Get rid of all annotated7 upstream transcription factor binding sites • Actually remove the sequence, don’t just insert the new promoter • Get rid of as many potential (i.e. based only on motif match) transcription factor regulation sites as possible But: • Avoid disrupting transcription initiation or termination of upstream genes Case 1: Upstream gene in same orientation as target gene MSG5 The synthetic pathway will be characterized to determine the effects of these modifications and to extend our understanding of pathway function. Assaying such characteristics as mating efficiency, recovery from pheromone-induced cell-cycle arrest and genome-wide transcription levels will allow us to understand the contributions made by wild-type transcriptional regulation to pathway function and mating program induction. Case 2: Upstream gene in opposite orientation as target gene FUS3 Intergenic region Intergenic region Upstream gene … STOP STE12 ATG …Target gene Upstream gene GTA ATG …Target gene TF binding sites Upstream gene … STOP FAR1 Pathway overview 1. Over what range of constitutive (i.e. not mediated by STE12) expression of single proteins does the pathway retain proper function ? 2. How robust is the pathway to the removal of multiple transcriptional feedback loops ? We will answer these questions by removing the STE12-mediated transcriptional feedback loops, both singly and in combination. Pathway genes that are STE12-responsive in the wild-type pathway will be placed under the control of promoters that allow us to vary their mRNA levels across a large range, and the response of the altered pathway will be characterized. The results of these experiments will also allow us to refine our computational model. Constitutive STE2 synthesis Pheromone-induced STE2 synthesis •Adaptation to downregulate pathway activity appropriately •Resistance to cell cycle arrest when exposed to pheromone outside G1 •Maintenance of specificity e.g. avoiding inappropriate activation of the filamentous growth pathway Active GPA1 (molecules/cell) STE2 (molecules/cell) 4000 3000 600 400 200 0 50 100 150 Time (min) 200 6000 6000 5500 5000 250 0 -50 300 0 50 100 150 Time (min) 200 250 5000 4500 4000 Constitutive STE2 synthesis Pheromone-induced STE2 synthesis 0 50 The functioning of the re-engineered pathway thus needs to be examined at multiple levels. We will examine pathway function by: • Monitoring the expression of a chromosomally-integrated copy of YFP driven off the pheromone-responsive PRM1 promoter, which will provide insight on the timecourse of signaling through the pathway • Generating dose-response curves of pheromone sensitivity, to characterize the impact on basal signaling • Examining the efficiency of recovery from cell-cycle arrest upon removal of pheromone, thereby measuring the extent to which pathway desensitization is affected • Generating genome-wide transcript measurements, to obtain a comprehensive picture of the molecular response induced by the altered pathway connectivity • Assaying mating efficiency, as a means of gauging the overall impact of the changes to the pathway References and Acknowledgements 300 We would like to thank members of the Endy and Knight labs, Dr. E. Fraenkel and Dr. A. van Oudenaarden at MIT for providing valuable feedback. Discussions with Dr. F. Winston of Harvard and Drs. K. Benjamin, A. Colman-Lerner, R. Yu and G. Pesce of the Molecular Sciences Institute also provided key insights. This work was funded by the Computational and Systems Biology Initiative at MIT. 3800 3600 3000 -50 Yeast cells react to pheromone on a wide range of timescales, from a response time on the order of seconds by the signaling cascade component of the pheromone response pathway, to the actual mating program, which can take several hours to complete. The pheromone response also produces a wide variety of phenotypes, from morphological changes such as the formation of a mating projection and cell-cycle arrest to the differential transcription of several hundred genes. We are interested in understanding the contribution to proper function of the yeast pheromone response pathway made by pheromone-mediated transcriptional induction of pathway components and regulators. In this poster, we describe our motivating questions and our approach to answering these questions, namely placing pathway genes under controllable promoters and characterizing the pathway’s response to removal of one or more transcriptional feedback loops. At present, we have constructed strains containing the STE2 and FAR1 genes under the control of constitutive promoters and are in the process of characterizing these strains. 1000 3500 Will decide how much to replace on instance-by-instance basis, taking into account factors like relevance of upstream gene to mating, higher stringency for putative regulatory sites based purely on motif matching etc Conclusions 800 100 150 Time (min) 200 250 3400 SST2 (molecules/cell) •Efficient induction of the cellular program required for mating in response to pheromone 5000 Active FUS3 (molecules/cell) •A level of basal activity that prevents inappropriate differentiation 6000 2000 Active STE12 (molecules/cell) Proper control of the pathway thus has several facets: New promoter ATG …Target gene Constitutive STE2 synthesis Pheromone-induced STE2 synthesis 1000 7000 0 -50 ? 1200 8000 Yeast can exist as haploid cells, in two “mating types” termed a and alpha, with mating between cells of opposite mating types resulting in a single diploid yeast cell. Mating between yeast cells is mediated by the pheromone response pathway, which is activated when a haploid cell senses the presence of pheromones emitted by cells of the opposite mating type. Activation of the pathway results in transient differentiation of the cell via cell cycle arrest in G1, the formation of a mating projection and differential transcription of several hundred genes1. If mating does not occur, or the stimulus is removed, the cells re-enter and proceed through the rest of the mitotic cell cycle. In addition to mediating the response to pheromone, the pathway also shares some molecular machinery with pathways activated under different conditions, such as the High Osmolarity Glycerol (HOG) pathway. Upstream gene GTA Characterizing the modified pathway Previous work has shown that constitutive overexpression of the genes upregulated by STE12 does not, in the majority of cases, lead to constitutive pathway activation or total loss of signaling upon exposure to pheromone. This argues that pathway function is robust in the face of above-basal constitutive levels of expression of single genes and single changes to the transcriptional architecture of the system (i.e. removal of one transcriptional feedback loop). In addition, a detailed computational model of the pathway (developed in our laboratory2) predicts pathway dynamics that are largely independent of transcriptional induction of certain genes, as illustrated below for the receptor STE2, a gene that is upregulated by a factor of five on exposure to pherome1. This naturally leads to two questions: 9000 ATG …Target gene A terminator built into the new promoter will cause proper transcription termination of the upstream gene, so we can replace as much as we need to of the upstream region. Are transcriptional feedback loops necessary for pathway function ? (K. Benjamin, Molecular Sciences Institute, unpublished) New promoter TF binding sites 4000 3000 2000 3000 (2) T. Thomson, unpublished 2800 (3) K. Benjamin, personal communication. 2600 2400 1000 300 (1) Roberts CJ et al, 2000. Science 287:873. 3200 0 -50 Constitutive STE2 synthesis Pheromone-induced STE2 synthesis 0 50 100 150 Time (min) 200 250 300 2000 -50 (4) Alper H, Fischer C, Nevoigt E, Stephanopoulos G, 2005. Proc Natl Acad Sci USA 102:12678 Constitutive STE2 synthesis Pheromone-induced STE2 synthesis 2200 0 50 100 150 Time (min) 200 250 300 (5) Urlinger S et al, 2000. Proc Natl Acad Sci USA 97:7963 (6) Becskei A, Kaufmann BB, van Oudenaarden A, 2005. Nat Genet 435:937 (7) Harbison CT et al, 2004. Nature 431:99