* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Synthesis and elongation of fatty acids
Basal metabolic rate wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Proteolysis wikipedia , lookup
Genetic code wikipedia , lookup
Point mutation wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Western blot wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Lipid signaling wikipedia , lookup
Citric acid cycle wikipedia , lookup
Peptide synthesis wikipedia , lookup
Specialized pro-resolving mediators wikipedia , lookup
Butyric acid wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Biosynthesis of doxorubicin wikipedia , lookup
Biosynthesis wikipedia , lookup
Synthesis and elongation of fatty acids
A molecular caliper mechanism for determining
very long-chain fatty acid length
Vladimir Denic and Jonathan S. Weissman (2007)
Cell 130, 663-677
February 28, 2008
Toon de Kroon
Membrane Enzymology
Reasons for choosing this topic/paper:
• Acyl chains: membrane building blocks
• Synthesis of fatty acids
• Specific functions of very long chain fatty acids
• Yeast as powerful model eukaryote (in lipid research)
• Functional reconstitution of membrane protein enzymes
Outline of introduction
Fatty acids
•Function
•Review of FA synthesis
general
Very long chain fatty acids
•Definition
•Function
•Synthesis: elongation
“Yeast tricks”
yeast
Fatty acids
• Essential in all organisms except archaea
• Constituents of membranes
• Posttranslational protein modification
(myristoylation, palmitoylation)
• Storage of chemical energy
(TAG, sterolesters)
Eukaryotic membrane lipids
Acyl chain composition determines
physicochemical membrane parameters:
{
acyl chain length
degree of unsaturation
membrane thickness
membrane fluidity
membrane curvature (PE, DAG)
which affect membrane protein function
molecular shape
PC
PI
PE confers negative curvature stress
PE
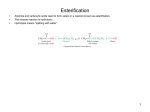
Synthesis of fatty acids
The formation of malonyl-CoA is the committed step
in fatty acid synthesis
acetyl-CoA carboxylase
(biotin)
Intermediates in fatty acid synthesis are attached to ACP
Fatty acid synthesis:
sequential addition
of two-carbon units
up to C16, by fatty
acid synthase (FAS),
localized in the cytosol
acyl-malonyl condensing enzyme
(β-ketoacyl synthase) KS/CE
β-ketoacyl reductase KR
β-hydroxyacyl dehydratase
DH
enoyl reductase ER
Stoichiometry of the synthesis of C16:0
8 acetyl CoA + 7 ATP + 14 NADPH + 6 H+
palmitate (C16:0) + 14 NADP+ + 8 CoA + 6 H2O + 7 ADP + 7 Pi
Animal fatty acid synthase: homodimer of single chains
(540 kDa)
AT, acetyltransferase
MT, malonyltransferase
KS/CE, condensing enzyme
KR, β-ketoacyl reductase
DH, dehydratase
ER, enoylreductase
TE, thioesterase
FAS in fungi: α6β6
2.6 MDa particle
Jenni et al., 2007
Domain organization and structure of the subunits of fungal FAS
Jenni et al., 2007
Reaction chambers of yeast FAS
pathway traversed by ACP
side view
ACP in modeled positions
at each active site
top view
Lomakin et al., 2007
Desaturation of fatty acids:
by electron transport chain in the ER
Phospholipid and acyl chain composition of wild type yeast
phospholipid class
phosphatidylcholine (PC)
phosphatidylethanolamine (PE)
phosphatidylinositol (PI)
phosphatidylserine (PS)
cardiolipin (CL)
mol%
41
26
18
9
5
fatty acid mol%
C14
C16:0
C16:1
C18:0
C18:1
C26:0
2
15
48
4
30
1
Very long chain fatty acids (VLCFA): > C18
Functions
in mammals:
- >C30: skin permeability barrier
- signaling: e.g. C20:4 (arachidonic acid)
- constituent of myelin
in plants:
- oils and waxes
in S. cerevisiae:
- stabilization highly curved membranes
- required for sphingolipid synthesis
- required for GPI-anchors
Morphological phenotype of yeast acc1 ts mutant at 37oC
(is not rescued by C16 supplementation)
•Separation nuclear inner and
outer membranes (asterisk)
•Formation of vesicle-like
structures (star)
•Arrow points to nuclear pore
0.6µm
Schneiter & Kohlwein, 1997
Models of highly curved membrane domains and their
possible stabilization by VLCFA-containing lipids
C26-PI
Schneiter et al., 2004
Sphingolipid metabolism in Saccharomyces cerevisiae
long chain bases (LCB)
Structure of GPI anchors of yeast: C26:0
Synthesis and elongation by
different enzyme complexes:
similar reactions
little sequence homology
Fatty acid elongation machinery in S. cerevisiae
Sur4p (Elo3p)
elongation up to C26
Fen1p (Elo2p)
elongation up to C22
∆elo2∆elo3: inviable
(Elo1p
elongation up to C18
elo1 mutants can’t use C12-14 as substituents for
endogenous fatty acid synthesis)
Ybr159wp:
Dehydratase:
Tsc13p:
β-ketoacylreductase: not essential
??
enoyl reductase: essential
Issues addressed by Denic and Weissman:
• Function of the Elops Fen2p and Sur4p in elongation
• Gene-enzyme relationships in FA elongation
• Assignment of the β-hydroxyacyldehydratase
• How is the length of the fatty acid determined?
to be presented by: Agon Hyseni
Tessa Quax
Bastiaan Bijl
Myrthe Braam
Yeast tricks
1. DAmP (decreased abundance by mRNA perturbation)
trick to include essential genes in large-scale
screens for genetic interactions (e.g. synthetic
lethal/sick screens):
the 3’-UTR (untranslated region) of the gene is disrupted by
insertion of an antibiotic resistance marker, destabilizing the
corresponding mRNA, yielding proteins under their natural
transcriptional regulation but at substantially reduced levels
2. The plasmid shuffle
Generation of a library of mutant alleles of an essential gene
leu2 ura3 trp1 yfg::LEU2
YFG1
URA3
TRP1
leu2 ura3 trp1 yfg::LEU2
yfg1-x
yfg1-x
TRP1
Selection for loss of URA3
plasmid on 5-FOA (ura+)
Only cells that have lost the wild type gene on the
URA3 plasmid can grow on the 5-FOA agar plate