* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Carbohydrates
Molecular evolution wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Western blot wikipedia , lookup
Gene expression wikipedia , lookup
Bottromycin wikipedia , lookup
Peptide synthesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
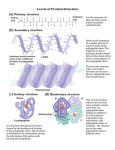
Protein moonlighting wikipedia , lookup
Phosphorylation wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Expanded genetic code wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Protein folding wikipedia , lookup
Genetic code wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Metalloprotein wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Point mutation wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Carbohydrates Sugar classification & STRUCTURE Sugars! = “Saccharides”: (“-OSE”) Monosaccharides Ex. Glucose, Fructose, Galactose Note: Glucose molecules (and others) form rings in aqueous solutions; not linear form that we may draw. Two forms of glucose = ᾳ & β forms (Geometric isomers) Dehydration synthesis of sugars form covalent bonds called Glycosidic Linkages. Disaccharides Ex. Maltose = glucose + glucose Sucrose = glucose + fructose Lactose = glucose = galactose 1 Polysaccharides (These are carbo’s Macromolecules) Ex. Starch = polymer of all glucose monomers Two types: 1. Amylose = unbranched 2. Amylopectin = branched Glycogen = polymer of all glucose monomers; more extensively branched than amylopectin. Chitin = branched Cellulose = unbranched 2 FUNCTION Function depends on structure = determined by its number and type of monosaccharides and by position of glycosidic linkages. Mono- & Disaccharide - 1st source of energy for most organisms - Carbon skeleton provides raw material for synthesis of other organics; ex., amino acids and lipids. Polysaccharides Energy storage 1. Starch = Plants 2. Glycogen = In liver and muscle of humans and other vertebrates Structural 1. Chitin = exoskeleton for insects and some marine animals. (shrimp, crab) 2. Cellulose = (all β glucose – every other glucose is upside down; forms straight chain. - Due to structure, chains can stack on top of one another = Strong! - Major component cell walls in plants and some bacteria. Cell Identification w/Protein ….more later 3 Lipids Fats & Oils, Phospholipids, Steroids, Waxes Hydrophobic based on structure; Oxygen scarce and no nitrogen = very little electronegativity = non-polar. 1 gram of fat stores twice as much energy than 1 gram of polysaccharide (Carb). WHY? Covalent bonds of long Hydrocarbon chains,…lots of energy in those long chains! Harder to break all those H-C bonds (vs. bonds in carbs that have oxygen at the bond). 1. Fats – not polymers, but are large STRUCTURE Composed of glycerol and fatty acids. Ex. Triglyceride = 3 fatty acid + 1 glycerol H-C covalent bonds in hydrocarbon chain = non-polar - Ester linkage = bond between hydroxyl group of glycerol and carboxyl group of fatty acid. 4 Fatty acids vary in length and in the number and locations of double bonds. - No double bonds = saturated fat (solid @ room temp) o Packs tightly - One or more double bonds (by removal of H) = unsaturated fat (liquid @ room temp.) Unsaturated cont… o In H-C chain, where a cis double bond occurs = kink in the chain = cannot pack close together = fluidity o In H-C chain, where a trans-double bond occurs = allow tighter packing, but can occur in both saturated and unsaturated fats. Contributes to heart disease. 5 FUNCTION Energy storage in animals cells. - Stored in adipose cells. Insulation Cushions vital organs 2. Phospholipids STRUCTURE Two fatty acids attached to glycerol (1 or 3 in fats) = “tails” = hydrophobic Functional group = phosphate = “head” = negative charge = hydrophilic - Charged or polar molecules can attach to phosphate (“head”) = variety of phospholipids Forms bilayer when in water. 6 FUNCTION Formation of cell membrane 1. Boundary for inside & outside cell… 2. Regulates cell’s internal environment = semi- or selectively permeable. 3. Steroids STRUCTURE Carbon skeleton made up of 4 fused rings. Differ in functional groups. - Example: Cholesterol & sex hormones FUNCTION Estrogen & Testosterone = Chemical messengers; coordinates cell activities of an organism. Cholesterol = Helps maintain the fluidity of the membrane Protein Remove the water from our cells and what’s left is mostly protein! Determines the structure and function of an organism. STRUCTURE 20 different amino acids - Functional groups - carboxyl group at one end (C terminus); amino group at the other end (N terminus). (Hence, …amino acid) 7 - General Structure: Amino acids differ in R group = can be non-polar, polar, or charged - R groups will give different properties to each amino acid; will help determine overall structure of a specific protein. - Covalent linkages = Peptide bonds = Carboxyl group of one amino acid (loses -OH) covalently bonds with (-H) of a different amino acid. 8 Protein Structure in Detail Polypeptide is to protein as is yarn is to a sweater. Amino acid sequence (number of and location of a.a) determines protein conformation. = chemical properties of Rgroups. Folding is reinforced by various types of bonds = 3-D conformation. Final conformation determines protein’s function. Almost always, the function of a protein depends on its ability to recognize and bond to another molecule. 4 levels of Protein Structure 1. Primary (1) = Unique sequence of a.a. (order & # of a.a.) = code in DNA - Shape = straight chain (polypeptide) - Covalent bond = peptide bond between a.a. 2. Secondary (2) - Shape o coils = alpha () helix o folds = beta () pleated sheet - H-bonds = between the a.a. backbones (of different polypeptides ~ 1). o N and O = electronegative. H of amino group in one polypeptide H-bonds with O of carboxyl group in a second polypeptide. 9 - Specifics of H-bonds a. ᾳ - helix = coil - H-bonding between every 4th a.a. - 3 non-polar a.a., then 1 charged a.a.,…and so on b. β pleated sheet - H-bonds on all a.a. between polypeptide backbones 3. Tertiary (3) – superimposed secondary structures;. - Shape = globular protein - Bonds = H-bonds, Van der Waals forces, ionic bonds, hydrophobic interactions, and disulfide bridges (covalent) bonds stabilize protein conformation. Interactions between R-Groups o H-bonds between polar R groups. o Hydrophobic interactions (due to presence of water) between non-polar R groups = cluster together; once in cluster held together by Van der Walls forces o Ionic bonds between charged R groups. 10 o Covalent bonds = disulfide bridges (S-S) = S of Sulfhydryl groups of different a.a. are brought close together in the folding of the protein for extra reinforcement of shape. Small part of hypothetical protein 4. Quaternary (4) – Aggregation of different 2° & 3° levels o One functional macromolecule o Bonds = various o Each subunit has a non-protein component - Ex. Hemoglobin = carries O2/CO2 in blood; contains iron - Ex. Collagen = helical subunits (2°) intertwined – connective tissue in skin, bone, tendon, ligaments Accounts for 40% of protein in humans 11 FUNCTION Speed up chemical reactions = ENZYMES! [S/F = Lock-N-Key Model] - Active site of enzyme has specific shape - Only fits specific substrate 12 Support Storage Transport Chemical messengers; coordinate organism’s activities on a macro and micro level. (hormones = Ex. Insulin) Response to chemical stimuli. Movement Defense against disease Nucleic Acids Two types : DNA & RNA The linear sequences of nucleotide in DNA are passes from parents to offspring - The sequences of bases determine a.a. sequence for a protein. 1 gene codes for 1 protein Organisms that share a greater proportion of DNA sequences are more closely related, evolutionarily; therefore, their protein structures are also similar. (E/R!) STRUCTURE Consists of polymers called polynucleotides. Monomers = Nucleotides - Nucleotide 1. Sugar (pentose); 13 - Deoxyribose = DNA (lack Oxygen on 2nd C in ring; hence = “Deoxy-“ - Ribose = RNA 2. Nitrogen Base: Two groups; differ in functional groups - N removes H+ from solution; hence = “BASE” - Note: Nucleoside = sugar & base 3. Phosphate – attaches to the 5´ carbon of the sugar. Note: Because the atoms in the sugars and the bases are both numbered, the sugars have a prime (´) after the number 14 Nulceotides are grouped based on size: - Purine = larger – 6 member ring fused to a 5 member ring of C & N. Adenine and Guanine - Pyrimidine = smaller – 6 member ring of C & N. Cytosine and Thymine (& Uracil) Building a polynucleotide: Adjacent nucleotides are covalently bonded = phosphodiester linkages; between the 3´ carbon of one nucleotide and the phosphate on the 5´ carbon on the next. = repeating pattern of sugars and phosphates down backbone. One end of the polynucleotide = phosphate attached to 5´ C = 5´ end Other end = hydroxyl on 3´ C = 3´ end Both strands in DNA are anti-parallel (2 one way streets running in the opposite directions) DNA is built from the 5´ to 3´ end! Sequence of bases attached to the sugars is unique to each gene. H-bonds hold the two anti-parallel strands together at the paired bases. 5´ ATCCGTA 3´ 3´ TAGGCAT 5´ Van der Waals forces hold together the stacked bases. 15 Van der Waals (between all base Pairs; only shown Once) Covalent bond; Phosphodiester linkage (Double)H-bond between A & T (triple)H-bond Between G & C FUNCTION Store genetic, hereditary information. Code to build proteins. 16