* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Supplemental Material 1 Simultaneous isolation of mRNA, miRNA
Protein moonlighting wikipedia , lookup
Genome evolution wikipedia , lookup
RNA silencing wikipedia , lookup
Epitranscriptome wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
RNA interference wikipedia , lookup
Western blot wikipedia , lookup
Gene therapy wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
List of types of proteins wikipedia , lookup
Gene expression wikipedia , lookup
Gene desert wikipedia , lookup
Gene nomenclature wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gene expression profiling wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
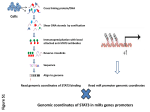
Supplemental Material 1 Simultaneous isolation of mRNA, miRNA and proteins from tissues mRNA and proteins were isolated from frozen tissue and select cell lines (wild-type SMCs and DICER-/-) using an AllPrep® DNA/RNA/Protein Mini Kit (QIAGEN AG, Basel, Switzerland) according to the published protocol. Enriched miRNAs were extracted and purified using the mirVana miRNA Isolation Kit (Ambion, Austin, TX), and their concentration and quality were determined with the NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Rockland, IL). The quality of RNAs was determined by the Agilent BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA). IPA To better understand the biological function and/or diseases that were most relevant to the data sets and facilitate understanding beyond a functional link to intracranial aneurysms (IA), ingenuity pathway analysis (IPA) was used to compare different types of cellular interactions, including gene-gene interactions and gene-miRNA interactions, with the genes analyzed above. IPA calculates the significance value of a given canonical pathway or biological function as the probability that the pathway or function is associated with the dataset by random chance. The dataset containing miRNA gene identifiers and corresponding fold changes was uploaded into the Pathway Studio, and each gene identifier was mapped to its corresponding gene object. The p value was calculated using the right-tailed Benjamini-Hochberg multiple testing correction with values of p < 0.05 considered significant. The “Core, Metabolomics Comparison Analysis and Path eExplorer” option of IPA was used to connect all of the identified proteins, mRNA and miRNAs associated with IA and its angiogenesis. Double immunofluorescence and confocal analyses Cells grown on coverslips were fixed with 4% formaldehyde for 20 min at room temperature, followed by 0.5% Triton X-100 treatment for 5 minutes and 3% BSA blocking. The slips were then incubated with the corresponding primary and secondary antibodies along with DAPI staining for visualization of nuclei. Fluorescence images were acquired with a Zeiss 710 META microscope. Plasmids 1 The 3’UTRs of MYOCD, ARHGEF12, FGF12, and ADCY5 were amplified by PCR from the genomic DNA of SMC and cloned into pcDNA3.1(+) according to standard protocols. The MYOCD 3’UTR was then subcloned into psiCHECK-2 using XhoI and NotI restriction sites. MYOCD 3‘UTR was also subcloned into the pMS2 vector for RIP analysis. Primer sequences were as follows: MYOCD, 3‘UTR-F GGACCTCACTCCGCCAAAT, MYOCD, 3’UTR-R TATTCCTCCACATCCCAC; TGTTAGTATATTCTTTTTCTTAATA, ARHGEF12, ARHGEF12, 3‘UTR1-F 3’UTR1-R ACCAGTCATTTAGCTTGAAAGAGGG; FGF12 3‘UTR2-F AAATGTCTGCAGTTCAAGAAAAGTT, FGF12, 3‘UTR2-R GTGTAAAGTGAGGGAAAC; and ADCY5, 3‘UTR-F CAAAGGCGAGATGATGAC, and ADCY5, 3’UTR-R CTTCACTTCTGGTCCCTACTCAGCT. Cycle Stretch A custom-made test apparatus was designed and manufactured for the application of cyclic tension capable of operating inside an incubator with a strain range of 0-25% and frequency range of 1-3 Hz. The tensile device consists of electrical and mechanical units. The electrical unit includes a PLC+HMI, a stepper motor driver, and a power supply. Data acquisition is performed using a computerized program. The mechanical unit contains a step motor, ballscrew, connector rod, an encoder and mobile and fixed grippers for stretching an elastic membrane according to a programmed pulsatile algorithm. The uniformity of applied strain was tested along the length of membrane by measuring the strains of stained locations on the stretched membrane. A maximum deviation of 0.5% of the measured strain value across the membrane ensured adequate manufacturing quality[2]. Reagents The following antibodies were used: anti-MYOCD antibody sc-33766 (Santa Cruz Biotechnology, Inc., Dallas, TX); anti-ARHGEF12 antibody 89-201-312 (ABNOVA, Taipei, Taiwan); anti-FGF12 antibody ab58157 (ABCAM, Cambridge, MA); anti-SM22 antibody sc-51442 (Santa Cruz Biotechnology, Inc.); and anti-ADCY5 antibody CAB-2602MH (Creative Biomart, Shirley, NY). The following siGENOME siRNA reagents were used: for nontargeting, 2 (siNC); MYOCD (siMYOCD); ARHGEF12(siARHGEF12); FGF12(siFGF12); ADCY5(siADCY5); and Dharmafect 1 (Dharmacon). The following 2 reagents and kits were used: Lipofectamine 2000, Trizol reagent, Dulbecco’s modified Eagle medium (DMEM), Opti-MEM reduced serum media, and fetal bovine serum (FBS) (Invitrogen Life Technologies, Carlsbad, CA); psiCHECK-2 vector and dual-luciferase reporter assay (Promega, Madison, WI); and RNeasy mini kit, DNeasy blood and tissue kit, and Qiaprep spin miniprep kit (Qiagen, Manchester, UK). IA model and survival analysis The left common carotid artery and the posterior branches of both renal arteries of 30 male 7-week-old C57BL/6J mice (200-300 g) were ligated to induce cerebral aneurysms. These procedures were performed with the mice under intraperitoneal zoletile anesthesia (30 mg/kg) with xylazine (10 mg/kg) and additional injections if necessary. After the operation, 1% normal saline was substituted for the drinking water to enhance the degree of hypertension. Then, the mice were intraperitoneally injected with 3 doses of 1-8 mg of tamoxifen (T5648, Sigma-Aldrich Co. LLC., St. Louis, MO) freshly dissolved in 100 μl corn oil (C8267, Sigma-Aldrich Co.) at 10 mg/ml, 20 mg/ml, 40 mg/ml or corn oil ehicle[3]. The subcutaneous injection site was sealed with a drop of Vetbond tissue adhesive (3M). Following tamoxifen administration, the mice were housed individually for 5-10 days before being analyzed for Cre-recombinase-mediated activity[4]. All animal care and experimental designs in this study complied with community standards on the care and use of laboratory animals. The survival analyses were performed on C57BL/6J mice IA models of wild-type MYOCD (n = 28) and MYOCD-/- (n = 13) mice, the high-expressing MYOCD (n = 13) or ceRNA group (ARHGEF12 n = 16; FGF12 n = 13; ADCY5 n = 12) and the low-expressing MYOCD (n = 15) or ceRNA group (ARHGEF12 n = 12; FGF12 n = 15, ADCY5 n = 16) according to the median within the wild-type group. Respectively, the survival analyses were performed on secondary hemorrhages of IA patients of the high-expressing MYOCD (n = 26) or ceRNA group (ARHGEF12 n = 29; FGF12 n = 23; ADCY5 n = 20) and the low-expressing MYOCD (n = 24) or ceRNA group (ARHGEF12 n = 21; FGF12 n = 27, ADCY5 n = 30) according to the median of gene expression levels, see also Fig. 7B, C. ceRNA criteria according to Tay et al [1]: We took a list of 5 miRs that were known from the literature to regulate MYOCD and they 3 generated a list of other, likely targets for these miRs using rna22. First they discard all predicted targets that have fewer than 2/5 miRs in common with MYOCD. Next, they filter these likely targets by a few other criteria to bolster confidence that some of the targets compete with MYOCD for miR binding. Gene X is predicted to be a ceRNA of MYOCD if: 1) Genes X and MYOCD have many miRs in common. This criteria is expressed as (the number of MYOCD miRs that are predicted to target gene X’s 3’UTR) / (the total number of miRs that target the MYOCD 3’UTR). The more miRs the two transcripts are predicted to share, the more this ratio approximates a value of 1. As the number of predicted, shared miRs between two transcripts increases, so does the likelihood that at least some of the miRs actually regulate both transcripts. Furthermore, if two transcripts are to exhibit a crosstalk phenomenon, then sharing more miRs means that the two transcripts have more ways they can regulate one another. 2) Shared miRs are predicted to target gene X over a short region. For a given miR, miR-n, gene X has a high ratio of (MREs for miR-n)/(distance in bases between left-most and right-most MREs of miR-n). This consideration arises because, longer 3’UTRs cause target prediction algorithms to return more spurious target predictions. This is approximated by the formula: E=K∗L1∗L2∗exp(−λS), where: • λ= A constant determined by the scoring system. • K = A constant determined by the database used. • L1 = The length of the 3’UTR. • L2 = The length of the MRE. • E = The mean of a P Poisson distribution of the number of matches between L1 and L2. As the length of 3’UTR under consideration (L1) increases, the higher the mean of the distribution becomes. Thus, target predictions across the entire length of a 3’UTR can be noisy. This criterion prioritizes targets with shorter islands because they are less likely to contain spurious predictions. 3) There is an even distribution of miR targets within target islands in gene X. These criteria are expressed as (distance between leftmost and rightmost MRE for miR-n)^2 / (sum of squared distances between miR-n MREs). This criterion does two things. First, it penalizes 4 targets where most but not all MREs aggregate in a small neighborhood. In this way, longer target islands are penalized because they may introduce spurious predictions. Second, it rewards instances that maximize the minimum distance between the same MREs. The MREs that are targeted by miR-n must be spaced out enough that miR-n+RISC can sit on the mRNA target without steric interference by another miR-n+RISC complex. By maximizing the minimum distance between MREs, we increase confidence that miR binding to all these MREs is possible simultaneously. 4) Fewest possible miRs give rise to a large number of MREs in gene X. This can be expressed as [(All MREs in transcript X for all miRs predicted to target X’s 3’UTR) - (# of miRs predicted to target X’s 3’UTR + 1)] / (MREs in X’s 3’UTR for all considered miRs). This criteria rewards transcripts for having a higher number of predicted MREs for each targeting miR. As the number of predicted MREs in target X for a particular miR increases, so does the likelihood that the miR actually regulates the target in at least one place. Once 1-4 above are calculated for all the miRs validated to regulate a gene of interest, the values can be multiplied out for every protein-coding transcript in the transcriptome to generate MuTaME scores. The transcriptome can then be rank-ordered based on the likelihood each gene functions as a ceRNA of your gene of interest. To review, the MuTaMe scoring approach takes validated regulatory miRs of a gene of interest and uses rna22 to generate lists of other genes likely regulated by those same miRs. This list is then rank-ordered by applying the criteria above which prioritize potential ceRNAs based on the number of miRs they share with a gene of interest, attributes of ceRNA target islands, and how likely miRs are to regulate ceRNA 3’UTRs. References: [1]. Tay, Y., et al., Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell, 2011. 147(2): p. 344-57. [2]. Ghazanfari, S., M. Tafazzoli-Shadpour and M.A. Shokrgozar, Effects of cyclic stretch on proliferation of mesenchymal stem cells and their differentiation to smooth muscle cells. Biochem Biophys Res Commun, 2009. 388(3): p. 601-5. [3]. Wicksteed, B., et al., Conditional gene targeting in mouse pancreatic ss-Cells: analysis of ectopic Cre transgene expression in the brain. Diabetes, 2010. 59(12): p. 3090-8. [4]. Wirth, A., et al., G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med, 2008. 14(1): p. 64-8. 5 6