* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download plasmid
Somatic cell nuclear transfer wikipedia , lookup
Gene prediction wikipedia , lookup
Gene therapy wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Restriction enzyme wikipedia , lookup
Deoxyribozyme wikipedia , lookup
DNA supercoil wikipedia , lookup
Community fingerprinting wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genetic engineering wikipedia , lookup
Designer baby wikipedia , lookup
Human cloning wikipedia , lookup
Expression vector wikipedia , lookup
DNA vaccination wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Molecular cloning wikipedia , lookup
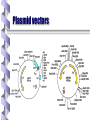
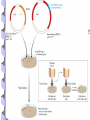
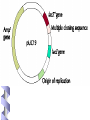
Fundamental of Biotechnology Plasmids Introduction The term plasmid was first introduced by the American molecular biologist Joshua Lederberg in 1952. Introduction Plasmid Self replicating, double Circular DNA molecule extrachromosomal (bacteria, yeast) stranded, entities in the cell Continue!! Size from less than 1kb to more than 500kb. (approx 0.1to 5% of the total DNA) Like the host-cell chromosomal DNA, plasmid DNA is duplicated before every cell division. at least one copy of the plasmid DNA is segregated to each daughter cell Continue!! # of identical plasmids within a single cell can range from one to even thousands (under some circumstances). Called high copy number plasmid (10-100 copies per host cell) Low copy number plasmid (1-4 copies per cell) Carry information for their own transfer (F Plasmid) Resistance to antibiotics (R Plasmid) Narrow and Broad Host Range Plasmids Due to the specificity of origin of replication: Narrow host range Plasmid: Replicate in only one spices of host cell. Broad host range Plasmid: less specific origin of replication Conjugative / Non conjugative Conjugative: self transmissible, tra (transfer) and mob (mobilising) regions carried on the plasmid. Non conjugative: not self transmissible by conjugative proficient plasmid if their mob region is functional. Continue!! Conjugative plasmid large= Low copy number Non conjugative small= high copy number Plasmid Cloning Vectors Plasmid cloning vectors have to be genetically engineered Naturally occurring plasmids often lacks important features such as Small size (>15kb decrease transfer in E. Coli) Choice of unique restriction site (to clone the DNA of interest) One or more selectable markers (to identify the recipient cells) MCS Plasmids cloning vector ORI Ab. Resis Three features of the plasmid cloning vectors: Multiple cloning site. The place where foreign DNA fragments can be inserted. An origin of replication: The replication origin is a specific DNA sequence of 50-100 base pairs that must be present in a plasmid for it to replicate. Host-cell enzymes bind to ORI, initiating replication of the circular DNA. A gene specifying resistance to an Antibiotic. This permits selective growth of the host cell. Most often used: Resistance to ampicillin, penicillin, tetracycline, kanamycin, and chloramphenicol. A plasmid vector for cloning 1. Contains an origin of replication, allowing for replication independent of host’s genome. 2. Contains Selective markers: Selection of cells containing a plasmid Contains a multiple cloning site (MCS) 3. Easy to be isolated from the host cell. Plasmid vectors Nomenclature Plasmid is designated by lower case such as p (stands for plasmid). Descriptive abbreviation some time e.g. pBR322 BR recognises the research work of F. Boliver and R. Rodriguez. 322 relevance to these workers. Plasmid vector pBR322 First plasmid vectors to be developed was pBR322 (Bolivar et al., 1977), constructed by ligating restriction fragments from three naturally occurring E. coli plasmids: R1, R6.5 and pMB1. The pBR322 plasmid is small (just 4361 bp) having the origin of replication, markers for double selection such as two antibiotics: Ampicillin and Tetracycline Continue!! pBR322 Restriction sites Recognition sequences for seven restriction enzymes, six of these sites lie within one or other of the genes for antibiotic resistance. Tetr (BamHI, HinIII and SalI), Ampr (Pst I, PvuI, Sca I) Ecor1 in non coding region. Cloning DNA Into a Plasmid Vector To reduce unwanted product, treatment with alkaline phosphate to remove 5 prim phosphate group from linear plasmid DNA pAT 153 Deletion derivatives of pBR322. Removal of two fragments (705bp) by using HaeII. 3 fold increase in copy number. Better than pBR322. Other plasmid cloning vectors pBR322 was a well conceived cloning vector but has few cloning sites selection procedure is time consuming pUC19 is a plasmid cloning vector created by Messing and co-workers in the University of California. p =for plasmid and UC represents the University of California. circular double stranded DNA and has 2,686 base pairs Continue!! pUC19 : recombinants, can be easily distinguished from the nonrecombinants based on colour differences of colonies on growth media. pUC18 is similar to pUC19, but possess different restriction sites. Component of pUC 19 It has one ampR gene (ampicillin resistance gene), lac Z gene of E. coli and lac I gene. The multiple cloning site (MCS) or (polylinker) region is split into the lac Z gene (codons 6-7 of lac Z are replaced by MCS) The multiple cloning site (e.g. EcorI, SacI, KpnI, XmaI, SmaI, BamhI, XbaI, SalI, HincII, AccI, BspMI, PstI, SphI, HindII) origin of replication from pBR322. Multiple Cloning Sites Transformation and Selection Transformed cell will grow b.c ampicillin resistance (ampR) gene. Transformed of interest can be distinguished by looking at the colour of the colony they make on agar media. Recombinants will be white, whereas non-recombinants will bed blue in colour. This is the most notable feature of pUC19. Mechanism In the presence of Isopropyl β-D-1-thiogalactopyranoside (IPTG) in growth medium, bacteria synthesise both fragments of the enzyme. Both the fragments can together hydrolyse X-gal (5-bromo-4-chloro3-indolyl- beta-D-galactopyranoside) and form blue colonies on media with X-gal. Insertion of foreign DNA into the MCS located within the lac Z gene causes insertional inactivation of this gene at the N-terminal fragment of beta-galactosidase. Continue!! Thus bacteria carrying recombinant plasmids in the MCS cannot hydrolyse X-gal, giving rise to white colonies, which can be distinguished on culture media from nonrecombinant cells, which are blue. Therefore the media used should contain ampicillin, IPTG, and X-gal. Antibiotics Commonly Used as Selective Agents Plasmid vectors advantages and disadvantages Advantages: • Small, easy to handle • Straightforward selection strategies • Useful for cloning small DNA fragments (< 10kbp) Disadvantages: • Less useful for cloning large DNA fragments (> 10kbp)