* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Loss of the GP46/M-2 surface membrane
Site-specific recombinase technology wikipedia , lookup
DNA barcoding wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genome (book) wikipedia , lookup
Gene nomenclature wikipedia , lookup
Pathogenomics wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Metagenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome evolution wikipedia , lookup
Designer baby wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene expression profiling wikipedia , lookup
Koinophilia wikipedia , lookup
Hybrid (biology) wikipedia , lookup
Molecular and Biochemical Parasitology. 50 (1992) 151 160
~;~ 1992 Elsevier Science Publishers B.V. All rights reserved. / 0166-6851/92/$05.00
15 l
MOLBIO 01650
Loss of the GP46/M-2 surface membrane glycoprotein gene family in the
Leishmania braziliensis complex
D i a n e M c M a h o n - P r a t t 1, Y a r a T r a u b - C s e k o 1"*, K e n t o n L. L o h m a n l'**,
D . D . R o g e r s 2 a n d S t e p h e n M. Beverley 2
I Yale University, Department of Epidemiology and Public' Health, New Haven, CT, U.S.A. and aDepartment of Biochemistry
and Molecular Pharmacology, Harvard School of Medicine, Boston, MA, U.S.A.
(Received l l February 1991; accepted 16 August 1991)
Immunization with the GP46/M-2 membrane glycoprotein of Leishmania amazonensis has been shown to induce a
protective immune response against infection. We have surveyed a variety of trypanosomatid species and genera for the
presence and expression of this gene family, information that will be relevant to future vaccine studies against leishmaniasis.
Molecular karyotype analysis revealed the presence of GP46/M-2 genes in all members of the Leishmania mexicana complex,
Leishmania major, Leishmania donovani, Leishmania tarentolae, and Crithidia.fasciculata. In contrast, DNAs from species of
the Leishmania braziliensis complex (L. braziliensis, Leishmania guyanensis, and Leishmania panamensis) failed to hybridize to
GP46/M-2 probes. Western blot analyses with several polyclonal antisera against the GP46/M-2 protein revealed protein
expression in L. major and L. donovani, but not L. panamensis or L. braziliensis. Phylogenetic analysis suggests that a loss of
the GP46A gene family occurred following separation of the L. braziliensis complex, prior to speciation events within this
complex. These data indicate that GP46/M-2 membrane glycoprotein may not be critical to parasite survival, but may play an
ancillary role during the developmental cycle.
Key words: Trypanosome; Molecular karyotype; Protective antigen; Protozoon
Introduction
The GP46/M-2 membrane protein of Leishmania amazonensis is a major surface component of the promastigote (insect vector)
developmental stage of the parasite [1 3],
constituting approximately 1-2% of the total
membrane protein. The GP46/M-2 protein has
Correspondence address." D. McMahon-Pratt, Department of
Epidemiology and Public Health, Yale University School of
Medicine, P.O. Box 3333, 60 College Street, New Haven, CT
06510, U.S.A.
Present addresses." *Fundacao Oswaldo Cruz, Department of
Molecular Biology, Rio de Janeiro, R.J., Brazil. **Southwest
Foundation for Biomedical Research. San Antonio, TX 78284.
U.S.A.
Abbreviations: FBS, fetal bovine serum: SDS-PAGE, sodium
dodecylsulphate polyacrylamide gel electrophoresis.
been demonstrated, in conjunction with certain
adjuvants, to elicit protective immunity against
infection caused by L. amazonensis [4]. Structural studies indicate that the GP46/M-2
molecule is a glycoprotein molecule [3] which
is attached to the surface membrane via a
phosphatidylinositol-linked lipid anchor rather
than protectively embedded within the lipid
bilayer [5]. Neither the lipid anchor nor
associated carbohydrate side chain appear to
be responsible for the overall stability of the
molecule [5]. Based on the amino terminal
sequence of the GP46/M-2 protein, the gene
encoding the protein has been cloned and
sequenced from L. amazonensis [6]. The D N A
sequence data are consistent with the known
biochemical and immunochemical features of
the molecule and predicts a repetitive sequence
(24 amino acids repeated 4 times) within the
amino terminal portion of the molecule which
152
constitutes approximately 22% of the total
mature protein: the carboxy-terminal domain
consists of proline-rich and cysteine-rich areas
of sequence, which likely accounts for the
stability of this portion of the molecule to
proteolytic digestion. The sequence of the
molecules, however, is unique and appears
not to be related to any other molecule
sequenced to date. We were interested to
determine the phylogenetic distribution of the
GP46/M-2 gene as well as the expression of the
protein within various members of the genus.
This information is relevant for future vaccine
studies against leishmaniasis and potentially
might provide insight to the function of the
molecule.
Current data suggest that the GP46/M-2
gene family of L. amazonensis is heterogeneous
and encodes a family of nonidentical proteins
(refs. 6 and 7; P.J. Langer, personal communication). The studies detailed in this paper
show that members of the GP46/M-2 family
occur and are expressed in species of the
Leis'hmania major, Leishmania donovani and
Leishmania mexicana complexes, but not in the
Leishmania braziliensis complex. Evolutionary
considerations suggest that the gene(s) may
have been deleted in the L. braziliensis
complex.
Materials and Methods
Parasite stocks and culture. The parasites
used in this study and the sources of original
stocks are listed in Table I. Organisms were
cultured in either Schneider's Drosophila
medium or Medium 199 containing 10 to
20% heat-inactivated fetal bovine serum
(FBS; Gibco or Hyclone) according to methods described [8,9].
Molecular karyotype analyses. Leishmania
chromosomes were prepared in agarose plugs
and stored at 4°C in 200 mM Tris/100 mM
EDTA, pH 8.0 (storage buffer) as described
[10]. Pulsed field gel electrophoresis (PFGE)
was performed using the contour-clamped
homogeneous electric field (CHEF) method
o f C h u et al. [11], using an apparatus similar to
that described by these authors or a Bio-Rad
Model C H E F - D R II. Electrophoresis was
performed in 0.5 x TBE buffer (45 mM
Tris/45 mM boric acid/l mM EDTA, pH 8.3)
at 4°C using a voltage gradient of 6 V cm ~ at
a pulse time of 70 s for 24 h followed by a pulse
time of 150 s for 24 h. The gels were blotted to
Gene-Screen Plus (Dupont) or Nytran (Schleicher and Schuell), and hybridized with radiolabelled probes as described [12], using a
hybridization temperature of 6 0 C in 2 x
SSPE (1 × SSPE is 130 mM NaCI/10 mM
NaPO4/I mM EDTA, pH 7.6). Post-hybridization washes were 2 x 15 min with 2 ×
SSPE, 60'~C, 2 x 15 min with 2 x SSPE, 0.5%
S D S a t 6 0 ~ C , a n d 2 x 5 m i n w i t h 0 . 1 x SSPE
at room temperature. Previous studies with a
variety of hybridization probes have shown
that these conditions yield specific hybridization among divergent sequences, while further
decreases in temperature yield extensive nonspecific hybridization (unpublished data). Size
markers were oligomers of lambda DNA
(Bethesda Research Laboratories). Restriction
enzyme digestion of chromosomes in agarose
plugs was performed as described [12]. Probes
used for hybridization are indicated in the
figure legends [6].
Gel electrophoresis and Western blot analyses. Polyacrylamide gel electrophoresis in the
presence of sodium dodecylsulfate (SDSPAGE), was performed using 10% polyacrylamide without prior reduction of the samples
[14]. Western blot analyses were performed as
previously described [3] according to the
methods of Towbin et al. [15], employing
isolated promastigote membrane preparations
[3] and using 0.2 /IM pore sized nitrocellulose
sheets (Schleicher and Schuell). GP46/M-2
protein was detected using either the monoclonal antibody, M-2 which is specific for the
protein of L. amazonensis [16] or polyclonal
antisera raised in mice immunized with either
wild type vaccinia (negative immune control
sera) or recombinant GP46/M-2A using two
different vaccinia expression vectors (D. Rodriguez, J. Rodriquez, L. Rivas, K.L.L., M.
153
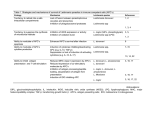
TABLE 1
Strains and species employed in this study
Stock
code
Species identification
Designation
Approximate
SignaP ~
GP46/M-2
chromosome size
Source b
MHOM/DO/00/450B
MHOM/MX/87/Mexl
MHOM/MX/87/Mex10
MNYC/BZ/62/M379
MHOM/PA/78/WR227
MHOM/BZ/82/BEL21
MHOM/BR/77/LTB0016
MPRO/BR/76/M4588
MHOM/BR/81/BOS-2
MORY/PA/79/GML3
MHOM/VE/74/PM-H3
MHOM/VE/75/HM76
500 kb
500 kb
500 kb
500 kb
8-900 kb
8 900 kb
8 900 kb
8 900 kb
8-900 kb
600 kb
5-600 kb
8 900 kb
i
i
i
a
c
e
f
a
j
a
a
a
MHOM/CO/82/CLO64
MHOM/CR/81/734
M H O M / P A / 7 4 / W R 120
MHOM/BR/75/M2903
MHOM/BR/79/LTB014
M H O M / B R / 7 0 / M 1"176
MHOM/BR/75/M4147
ND d
e
L. rnexicana complex
450B
Mex I
Mexl0
Lll c
WR227
BEL21
LTB0016
M4588
BOS-2
M6331
PM-H3 c
M5903
L.
L.
L.
L.
L.
L.
L.
L.
L.
L.
L.
L.
mexlcana complex
mexlcana
mexlcana
mexlcana
mexlcana
mexlcana
amazonensis
amazonensis
amazonensis
aristedesi
venezuelensis
garnhami
L. braziliensis complex
CLO64
L. panamensts
L. panamensts
CR734-81
WRI20
L. panamensts
M2903 c
L. braziliensis
L. braziliensis
LTB008
M1176
L. guyanensts
M4147 ~
L. guyanensts
IM350
L. guyanensts
Other Leishmania species
LRC-L38
L. mq]or
252
L. mq/or
LV9 c
L. donovani
LTC-I
L. tarentolae
Other genera
CFC- 1
C. jasciculata
LV88
E. monterogeii
Peru
T. cruzi
Moderate
Moderate
Moderate
Moderate
V. Strong
V. Strong
V. Strong
V. Strong
V. Strong
Strong
Moderate
V. Strong
ND
IUMB/BR/81/IM350
RHO/SU/XX/LRC-L38
MHOM/IR/83/LT252
MHOM/ET/67/L82
LTC-I
CFC- 1
LV88
PERU
n
k
c
a
b
a
a
m
ND
900 kb
800 kb
900 kb
ND
V. Strong
Strong
Strong
1100 kb
Moderate
~This characterizes the intensity of hybridization. As discussed in the text, these experiments do not determine whether the
variability observed arises from sequence divergence or changes in gene copy number.
bSources of strains were as follows: (a) Drs. J.J. Shaw and R. Lainson; (b) Dr. P. Marsden; (c) Dr. L. Hendricks; (d) Dr. C.C.
Wang; (e) Dr. D. Evans; (f) Dr. M. Hommel; (g) Dr. L. Simpson; (h) Dr. S. Mesnick; (i) Dr. F. Neva; (j) Dr. D. McMahonPratt; (k) Dr. L. Mata: (1) Dr. M. Chance; (m) Dr. J. Arias; (n) Dr. R. Tesh; (o) Dr. L. Schnur.
CWorld Health Organization Leishmania reference strains.
aND, not determined.
e( ) indicates no detectable signal.
Esteban and D.McM.-P. in preparation). The
GP46/M-2 polyclonal antisera recognized the
native GP46/M-2 of L. amazonensis and
previously characterized major proteolytic
fragments of the protein [3]. Antigens bound
by the monoclonal antibody or the mouse antiGP46/M-2 serum were identified with the use
of [125I]radioiodinated rabbit F(ab'2) antimouse immunoglobulin (2 x 105 cpm ml-1;
spec. act. 10 fCi/fg). Blots were dried and
exposed at - 7 0 ° C for autoradiography.
Results
Chromosomal distribution of the GP46/M-2
gene within the L. mexicana complex. Chromosomes of 12 members of the L. mexicana
complex were separated by pulsed field electrophoresis and examined by Southern blotting
with a GP46A probe. GP46/M-2 hybridization
was observed in all species tested, although 3
types of hybridization pattern were obtained
154
/
•
~. . . . . .
+¢
,y ,y,7,y
7,
~
•
I
I
I
CD
0
STRAIN
I
I
I
I
I
I
I
I
I
~0
~
m,
I
I
]
compression-zone
kb
9OO
80O
7OO
600
50O
400
300
200
I00
50
Fig. 1. Molecular karyotype analysis of GP46/M-2 genes in
the L. mexieana species complex. The experimental
conditions are as indicated in Materials and Methods.
Shown are the autoradiographic results from Southern blot
experiments of electrophoretically separated Leishmania
chromosomes and a radiolabeled GP46/M-2 probe. The
BamHl-Sphl restriction fragment of the L. amazonensLs'
GP46A gene was utilized, which contains all but the last 200
bp of the GP46A/M-2 coding region [I 3]. Molecular weight
markers (kb) are as indicated.
(Fig. 1; Table 1). In 6 lines (L. mexicana
WR227, Bel21: all three L. amazonensis and
Leishmania garnhami), strong hybridization
was observed to chromosomes of 800 950 kb
(Pattern 1), one chromosome in L. mexicana
WR227 and L. garnhami and 2 chromosomes
in each of the remaining 4 stocks. Hybridization to the sample loading well was also
evident. In Leishmania aristedesi somewhat
less intense hybridization was observed to a
single 600-kb chromosome as well as to the
sample well (Pattern 2; Fig. 1). In 4 lines of L.
mexicana (450B, MEX1, MEXI0, LI1) and
Leishmania venezuelensis, moderate hybridization was observed to a single 500 600 kb
chromosome (Pattern 3; Fig. 1). These data
revealed a complex karyotypic distribution of
GP46/M-2 within this species complex, although they do not determine whether changes
in copy number or sequence divergence are
responsible.
Chromosomal distribution of GP46/M-2 genes
in other species complexes and genera. Chromosomes from representatives of three other
complexes of pathogenic Leishmania ( L. ma/or.
L. donovani, L. guyanensis), the lizard parasite
Leishmania
tarentolae, and
the genera
Crithidia, Endotrvpanum and Trypanosoma
(Trypanosoma cruzi) were separated by pulsed
field electrophoresis and examined by Southern blotting with the GP46/M-2 probe (Fig.
2A,B). Examination of the ethidium bromide
stained gel (Fig. 2C) and hybridization with a
Leishmania ribosomal D N A probe (data not
shown) indicated comparable amounts of
D N A in all samples.
L. ma/or and L. donovani chromosomes of
930 and 850 kb exhibited strong hybridization
comparable to that seen in L. amazonensis
LTB0016. A 950-kb chromosome in L,
tarentolae showed moderate hybridization
comparable to that seen with some lines of L.
mexicana and L. t'enezuelensis, and a l l00-kb
chromosome in Crithidia hybridized weakly
(Fig. 2A,B; Fig. 1; Table I). As mentioned
earlier, these experiments do not determine
whether the variability observed arises from
sequence divergence or changes in gene copy
number. Finally, faint hybridization to 2 3
chromosomes in both Endoto'panum and T.
cruel was also observed, although the hybridization intensity in these species was only
slightly above background and may not be
significant (Fig. 2B).
In support of the hybridization data, GP46/
M-2-related sequences have been recently
isolated from molecular recombinant libraries
of both L. mq/or (S. Dasgupta, P. Murray,
personal communications)
and
Crithidia
(D.McM.-P., unpublished data).
Surprisingly, in L. guyanensis, no convincing
hybridization was observed, other than a faint
nonspecific hybridization to many other
chromosomes that was also evident in the
155
A
i
l
i
I
I
E
[
I
I
]
I
1
I
I
I
I
r
i
i
~
I
r
I
r
-compression
zone
)ression
kb
-
750
-
500
-
250
- 50
Fig. 2. Molecular karyotype analysis of GP46/M-2 genes in various genera of kinetoplastid flagellates. Southern
was performed as described in Fig. 1. The specific strains were: L. amazonensis (LTB0016), L. guyanensis
fasciculata (CFC-I), Endotrypanum monterogei (LV89), T. cruzi, L.-major (LT252), L. donovani (LV9) and
(M5903). Panels A and B differ only in the length of exposure of the hybridized blot to the film. In Panel C
bromide stained agarose gel is shown. Molecular weight markers (kb) are as indicated.
other species (Fig. 2B). This suggested that the
GP46/M-2 gene was absent in this species. To
confirm this, chromosomes were prepared
from other members of the L. braziliensis
complex, spanning a wide geographical range
and including representative strains of all 3
major recognized species, L. guyanensis, Leishmania panamensis and L. braziliensis (Fig. 3;
Table I). A long exposure of a blot of
separated chromosomes probed with GP46/
M-2 showed no hybridization above background to any of these species, although
massive hybridization was seen to the positive
controls L. major and L. amazonensis (Fig. 3,
right-hand panel; the faint pattern seen is
comparable to that observed with nonspecific
control hybridization probes such as pBR322
in other experiments). Examination of the
ethidium bromide-stained gel (Fig. 3, lefthand panel) and hybridization with an L.
major ribosomal R N A probe showed that
comparable amounts of D N A were present
on this blot (data not shown). Thus far, we
have not been able to provide evidence for the
GP46/M-2-related
sequences in the L.
braziliensis complex.
blot analysis
(M4147), C.
L. garnhami
the ethidium
Organization of GP46/M-2 genes in L. amazonensis, L. major and L. donovani. GP46/M-2
is known to be encoded as a family of nonidentical genes in L. amazonensis [6,7]. We
examined the structure of the GP46/M-2 gene
family in L. amazonensis LTB0016, which
contains two intensely hybridizing chromosomes (Pattern 1), by Southern blotting of
total D N A digested with infrequently cutting
restriction enzymes lacking sites in the GP46A
gene (Fig. 4A). Digestion with SpeI gave two
similarly intense fragments of 590 and 440 kb
(Fig. 4A), while Dral gave two fragments of
500 and 350 kb; digests with AseI, SspI, and
XbaI also yielded two comparably large
fragments differing in size by about 150 kb
(data not shown). In contrast, NdeI digestion
yielded at least 6 fragments, ranging from 8 to
l l 0 kb (Fig. 4A). Assuming the absence of
comigrating fragments, they total approximately 280 kb. Digestions with more frequently cutting enzymes similarly yielded multiple
differently sized fragments, however in no case
was a pattern indicative of a single tandemly
repeating repetitive array obtained (Fig. 4 and
data not shown; see also ref. 6).
156
/
/
/
I
I
/
;
(
I
i:,,
I
2
I
i
I
[
i
I
I
I
I
]
I
I
r :/U,
I
[
I
I
I
I
compression
zone
Fig. 3. Molecular karyotype analysis of GP46/M-2 genes in the L. braziliensiscomplex• The experimental conditions are those
indicated in Fig. 1, except that CHEF electrophoresis was performed using a pulse time orS0 s t\)r 18 h followed by a pulse
time of 160 s for 24 h. In the panel on the left is shown the ethidium bromide-stained agarose gel. On the right are shown the
autoradiographic results from the Southern blot analyses of the gel. The film was deliberately overexposed to emphasize the
lack of specific hybridization to chromosomes of the L. hraziliensiscomplex. Molecular weight markers (kb) are as indicated•
The data with the 5 infrequently cutting
enzymes provide evidence for 2 clusters o f
G P 4 6 / M - 2 genes in L. amazonensis LTB0016.
Interestingly, the two G P 4 6 / M - 2 c h r o m o s o m e s
differ by a p p r o x i m a t e l y 150 kb, as do the 2
large fragments with the 5 infrequent cutters.
This suggests that each G P 4 6 / M - 2 cluster is
located on a differently sized c h r o m o s o m e .
The simplest explanation is that an expansion
o f one G P 4 6 cluster occurred on one c h r o m o some h o m o l o g , in contrast to isolates such as
L. mexicana W R 2 2 7 and L l l which possess a
single G P 4 6 / M - 2 hybridizing c h r o m o s o m e
(Fig. 1). H o m o l o g o u s c h r o m o s o m e s differing
in size have been r e p o r t e d in Leishmania
[13,17 21] and t r y p a n o s o m e s [22 25].
The organization o f the G P 4 6 / M - 2 genes in
L. major and L. donovani was examined by
S o u t h e r n blotting with G P 4 6 / M - 2 probes after
digestion with infrequently cutting enzymes.
As observed with L. amazonensis, with some
enzymes one or two large fragments were
obtained, while with others a n u m b e r o f
smaller fragments were obtained (Fig. 4B,C).
With no e n z y m e was evidence o f a uniform
tandemly repeated array obtained (data not
shown). These data indicate that as for L.
amazonensis, G P 4 6 / M - 2 genes are organized in
a cluster o f nonidentical genes in these species.
Western blot analyses ji~r G P46/ M-2 expression
in Leishrnania species. The expression o f the
G P 4 6 / M - 2 protein in p r o m a s t i g o t e m e m b r a n e
preparations was examined by Western blol
analyses, using two polyclonal murine antisera
directed against the G P 4 6 A protein produced
by 2 different vaccinia vectors. Crossreactive
molecules were recognized by the polyclonal
157
A
B
L. amozonensls
C
L. m o j o r
L. donovoni
Mr
Mr
Mr
(kb)
(kb)
40O --
400 --
500 -
500 200
200 -
I O O
ioo
-
50
z&
-
L..--
"~9.4
5 0 --
2 5 1 -94--
6.6--
66--
4.4
....
0r}
Z
d3
U0
6.6
Z
Z
~
Z
"T"
C3
Fig. 4. Southern blot analysis of O P 4 6 / M - 2 genes in L. mqjor (LT252), L. donovani (LV9), and L. mexicana (M379).
C h r o m o s o m a l D N A s were digested with the indicated enzymes as detailed in Materials and Methods and resolved by C H E F
electrophoresis for 20 h using a 5 to 50 s ramp at 200 V. Gels w e r e s u b j e c t e d to Southern blot analysis with the HindlII-Sphl
fragment of the GP46A gene [6]. Identical results were obtained using the SalI-Hindlll fragment of the GP46A gene (not
shown). Molecular weight markers (kb) are as indicated for each panel.
L. panamensis
L. braziliensis
L. donovani
L. major
L. amazonensis
--
92
--68
--43
--30
......
1
2
3
,
....
4
5
--14
6
1
2
3
4
5
6
1 2 3 4 5
1234
5
12345
Fig. 5. Shown are the autoradiographic results from Western blot analysis of GP46/M-2 expression in 5 species of Leis'hmania
Membrane fractions isolated from L. amazonensis (LTB0016), L. mqjor (LRC-L38), L. donovani (LV9), L. braziliensis (M2903)
and L. panamensis (CLO64) were used as the source of antigen as described in Materials and Methods. In the case of L. major,
L. donovani and L. amazonensis, the murine antisera employed included: pre-immune/normal mouse sera (lanes 1); antivaccinia (wild type) control sera (lanes 2); anti-GP46/M-2-vaccinia recombinant sera (wild type) (lanes 3); anti-GP46/M-2vaccinia recombinant scra (mutant vector) (lanes 4); monoclonal antibody M-2 (specific for L. amazonensis) (lanes 5). For L.
braziliensis and L. panamensis the murine antisera employed included: pre-immune/normal mouse sera (lanes 1); anti-GP46/M2-vaccinia recombinant sera (wild type) (lanes 2); anti-GP46/M-2-vaccinia recombinant sera (mutant vector) (lanes 3);
monoclonal antibody M-2 (lanes 4); monoclonal antibody B-21, specific for L. braziliensis and L. panamensis (lanes 5);
monoclonal antibody L-l, cross-reactive for all Leishmania species (lanes 5). Molecular weight markers (kDa) are as indicated.
158
GP46/M-2 antisera in L. major and L.
donovani. Although positive control monoclonal antibodies (Fig. 5, lanes 5 and 6) gave
significant reactions under these experimental
conditions with the L. panamensis and L.
braziliensis membrane preparations, no crossreactions were observed with the GP46/M-2
polyclonal sera (Fig. 5, lanes 2 and 3). Similar
results were obtained in other Western blot
experiments employing membrane preparations of L. guyanensis (data not shown). As
expected, the L. amazonensis GP46-specific
monoclonal antibody M-2 only identified
GP46/M-2 in this species and not in either L.
ma/or or L. donovani (Fig. 6) [3,16]. Proteolytic
degradation products characterized previously
were observed in L. amazonensis [3], and it is
possible that the smaller 40 kDa band evident
in L. donovani and L. major arises in a similar
manner. The observation that polyclonal GP46
antisera recognizes proteolytic fragments suggests that even a truncated or partially
degraded L. panamensis or L. braziliensis
protein could have been detected if present.
Similar results were obtained in other experiments employing a rabbit antiserum against
native GP46/M-2 isolated from L. amazonensis
(data not shown). Thus, no evidence for
expression of a GP46/M-2-related protein has
been obtained in either L. braziliensis complex
promastigotes, the stage in which expression is
maximal in other species.
Discussion
We have surveyed the evolutionary distribution of the GP46/M-2 gene family in trypanosomatid protozoa, using a coding region probe
and conditions of relaxed hybridization stringency. These studies revealed the clear presence
of GP46/M-2-related genes in 3 species complexes of Leishmania pathogenic to humans (L.
mexicana, L. tropica, L. donovani), the lizard
parasite L. tarentolae, and the monogenetic
insect parasite Crithidia jasciculata. It is also
possible that sequences related to GP46/M-2
occur in Endotwpanum and Twpanosoma
cruzi, although additional studies will be
required to confirm this. In all species of
Leishmania GP46/M-2 genes are restricted to
one or two chromosomes, containing a cluster
with multiple nonidentical genes.
Within the L. mexicana species complex, at
least three different patterns for the chromosomal distribution of GP46/M-2 sequences
were found. Association of these patterns
with the species classification of each isolate
did not always reveal consistency; for example,
members of L. mexicana exhibited either
pattern 1 or 3. As both the structure of the
GP46/M-2 locus within this complex and the
evolutionary relationships amongst the isolates
examined become better understood, it may be
possible to interpret the evolution of the GP46/
M-2 karyotype in the future.
Unlike the other Leishmania tested, GP46/
M-2-related sequences could not be detected in
6 members of the L. braziliensis species
complex, isolated from a wide geographical
area and including the 3 recognized species.
These findings were supported by studies of
GP46/M-2 protein expression: GP46/M-2
expression was not found in promastigotes of
L. hraziliensis or L. panamensis, while GP46/
M-2 protein was readily detected in 1,.
donovani and L. ma/'or in addition to L.
amazonensis. These studies suggest that the
absence of GP46/M-2 sequences may be a
useful marker for members of the L.
hraziliensis complex.
Phylogenetic distribution of GP46/M-2. Phylogenetic methods were employed to analyze
the evolutionary distribution of GP46/M-2 in
Leishmania. Organismal [26] and molecular
evolutionary comparisons of nuclear DNA
small subunit ribosomal R N A sequences
indicate that CrithMia is an evolutionary
outgroup of all pathogenic Leishmania as well
as L. tarentolae (K. Nelson and S.M.B., in
preparation). As discussed above, current
molecular data suggest that Crithidia and
possibly more distant trypanosomatids possess members of the GP46/M-2 gene family.
The most parsimonious explanation is that a
single change was responsible for the absence
of GP46/M-2 sequences in the L. hraziliensis
159
complex, occurring after the separation of this
lineage from other Leishmania but prior to
speciation within this complex.
The simplest model that could account for
the lack of GP46/M-2 in the L. braziliensis
complex is a chromosomal deletion. Since
GP46/M-2 genes are organized as a gene
cluster in other Leishmania, this could readily
be accomplished in a single genetic event.
Chromosomal changes including expansion
and contraction of repetitive gene families
have been shown or postulated in numerous
isolates of Leishmania [13,17 21], although in
no instance has complete loss of a gene family
been reported. Alternatively, it is possible that
GP46/M-2 has undergone rapid evolution in
the L. braziliensis complex, sufficiently so that
the available antibody and hybridization
probes no longer recognize this protein or
gene. Although all gene family members would
have to undergo accelerated evolution, mechanisms such as concerted evolution [27]
could allow all members to evolve in parallel.
Although current data do not allow us to
choose between the deletion or accelerated
divergence models, we currently favor the
deletion model. In any event, it is clear that
the GP46/M-2 gene family shows a greater
variability in evolutionary distribution than
any other leishmanial protein antigen characterized thus far. For example, the surface
antigen gp63 is present in all Leishmania
species complexes and Crithidia [28,29]. This
suggests that GP46/M-2 may be associated
with features of the parasite life cycle that vary
among different species complexes. Unfortunately, the predicted protein sequence of a
GP46/M-2 gene from L. amazonensis does not
reveal affinity to any known protein that could
suggest a potential function [6].
It is known that GP46/M-2 is maximally
expressed in the promastigote stage within the
sand fly digestive tract (refs. 1, 2 and 30; and
D.McM.-P. et al., in preparation). Interestingly, one feature that distinguishes the L.
braziliensis complex from other Leishmania is
that these promastigotes are primarily found
within the hindgut, whereas promastigotes of
the other pathogenic species complexes pri-
marily reside within the midgut [26,31].
Conflicting data have been reported with L.
tarentolae [31], although one group has
reported midgut development in Phlebotomus
papatasii [32]. This group also reported midgut
development for C. fasciculata, although this
organism is normally carried by mosquitoes
[33]. These observations raise the possibility
that GP46/M-2 may affect parasite growth
within the midgut of the fly.
In one sense, members of the L. braziliensis
complex constitute evolutionarily derived
GP46/M-2 'null' mutants. The advent of
methods for stable DNA transfection of
pathogenic Leishmania [9] will permit exploitation of these 'mutants' in genetic tests of GP46/
M-2 function. We have developed a molecular
construct capable of directing GP46/M-2
expression, and succesfully introduced it into
L. panamensis, a member of the L. braziliensis
species complex [7,34]. The phenotype of these
genetically modified parasites during the
infectious cycle in the sand fly and macrophages may provide important clues about the
biological role of GP46/M-2 in the future.
Acknowledgements
The authors would like to thank Ms.
Kelledy Manson for excellent technical assistance. This work was supported by grants from
the National Institutes of Health AI-23004
(D.McM.-P) and AI-21903 (SMB). Yara
Traub-Cseko was supported by a CAPES/
Fulbright Fellowship.
References
1 McMahon-Pratt, D., Jaffe, C.L., Kahl, L., Langer, P.,
Lohman, K., Pan, A. and Rivas, L. (1987) Characterization of developmentally regulated molecules of
Leishmania. NATO Workshop: Host-Parasite Molecular Recognition and Interactions, HI I, 123 136.
2 McMahon-Pratt, D. and David, J.R. (1982) Monoclonal antibodies recognizing determinants specific for
the promastigote stage of Leishrnania m e x i c a n a
amazonensis. Mol. Biochem. Parasitol. 6, 317 327.
3 Kahl, L.P. and McMahon-Pratt, D. (1987) Structural
and antigenic characterization of a species- and
160
promastigote-specific Le&hmania mexk ana amazonensis"
membrane protein. J. Immunol. 138, 1587 1595.
4 Champsi, J. and McMahon-Pratt, D. (1988) Membrane
g l y c o p r o t e i n M-2 p r o t e c t s a g a i n s t Leishmania
amazonensis infection. Infect. Immun. 52, 3272 3279.
5 Rivas, L., Kahl, L., Manson, K. and McMahon-Pratt,
D. (1991) Biochemical characterization of the protective
m e m b r a n e protein, G P 4 6 / M - 2 of Leishmania
amazonensis. Mol. Biochem. Parasitol. 47, 235 244.
6 Lohman, K.L., Langer, P.J. and McMahon-Pratt, D.
(1990) Molecular cloning and characterization of the
immunologically protective surface glycoprotein GP6/
M-2 of Leishmania amazonensis. Proe. Natl. Acad. Sci.
USA 87, 8393 8397.
7 LeBowitz. J.H., Coburn, C.M., McMahon-Pratt, D.
and Beverley, S.M. (1990) Development of a stable
Leishmania expression vector and application to the
study of parasite surface antigen genes. Proc. Natl.
Acad. Sci. USA 87, 9736 9740.
8 Hendricks, L.D. and Hajduk, M.E. (1978) Haemoflagellates: commercially available liquid media for rapid
cultivation. Parasitology 76, 309 316.
9 Kapler, G.M., Coburn, C.M. and Beverley, S.M. (1990)
Transfection of the human parasite Leishmania delineates a 30 kb region sufficient for extrachromosomal
replication and expression. Mol. Cell. Biol. 10, 1084
1094.
10 Beverley, S.M. (1988) Characterization of the 'unusual"
mobility of large circular DNAs in pulsed field-gradient
electrophoresis. Nucleic Acids Res. 16, 925 938.
11 Chu, G., Vollrath, D. and Davis, R.W. (1986)
Separation of large D N A molecules by contourclamped homogeneous electric fields. Science 234,
1582 1585.
12 Beverley, S.M., lsmach, R.B. and McMahon-Pratt, 1).
(1987) Evolution of the genus Leishmania as revealed by
comparisons of nuclear D N A restriction fragment
patterns. Proc. Natl. Acad. Sci. USA 84, 484 488.
13 lowmnisci, D.M. and Beverley, S.M. (1989) Structural
alterations of chromosome 2 in Leishmania major as
evidence for diploidy, including spontaneous amplification of the mini-exon array. Mol. Biochem. Parasitol.
34, 177 188.
14 Laemmli, U.K. (1970) Cleavage of structural proteins
during the assembly of the head of bacteriophage T4.
Nature 227, 680 685.
15 Towbin, H., Staehlin, T. and G o r d o n , J. (1979)
Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some
applications. Proc. Natl. Acad. Sci. USA 76, 4350 4354.
16 Grimaldi Jr., G., David. J.R. and McMahon-Pratt, D.
(1987) Identification and distribution of New World
Leishmania species characterized by serodeme analysis
employing monoc[onal antibodies. Am. J. Trop. Med.
Hyg. 36, 270 287.
17 Scholler, J.K., Reed, S.G. and Stuart, K. (1986)
Molecular karyotype of specics and subspecies of
Leishmania. Mol. Biochem. Parasitol. 20, 279 293.
18 Bastien, P., Blaineau, C., Taminh, M., Rioux, J.A.,
Roizes, G. and Pages, M. (1990) Interclonal variations
in molecular karyotype in Leishmania in/antum imply a
"mosaic' strain structure. Mol. Biochem. Parasitol. 40,
53 62.
19 Samaras, N. and Spithill, T.W. (1987) Molecular
karyotype of five species of Le&hmania and analysis
of gene locations and chromosomal rearrangements.
Mol. Biochem. Parasitol. 25, 279 291.
20 Blaineau, C., Bastien, P., Rioux, J.-A., Roizds, G. and
Pages, M. (1991) Long-range restriction maps of sizevariable homologous chromosomes in Leishmania
il{/~Jntum. Mol. Biochem. Parasitol. 46, 293 302.
21 Bishop, R.P. (1990) Extensive homologies between
Leis'hmania donovani chromosomes of markedly different size. Mol. Biochem. Parasitol. 38, I 12.
22 Gibson, W.C. and Borst, P. (1986) Size-fractionation of
the s m a l l c h r o m o s o m e s of Trypanozoon and
Nannamonas by pulse field gel electrophoresis. Mol.
Biochem. Parasitol. 18, 127 140.
23 Gibson. W.C. and Miles, M.A. (1986)The karyotypc
and ploidy of Trypanosoma cruzi. EMBO J. 5. 1299
1305.
24 Henriksson. J., /i, slund, L., Macina, R.A.. de Cazzulo,
B.M.F., Cazzulo, J.J., Frasch, A.C.C. and Pettersson,
U. (1990) Chromosomal localization of seven cloned
antigen genes provides evidence of diploidy and further
demonstration of karyotype variability in Trypanosoma
cruzi. Mol. Biochem. Parasitol. 42, 213 224.
25 Gottesdiener, K., Garcia-Anoveros, J.. Lee, M.G.-S.
and Van der Plocg, L.H.T. (1990) C h r o m o s o m e
organization of the protozoan Tr)'panosoma hrucei.
Mol. Cell. Biol. 10, 6079 6083.
26 Lainson, R. and Shaw, J.J. (1987) Evolution, classification and geographical distribution of Leishmania. In:
The Leishmaniases in Biology and Medicine: Biology
and Epidemiology, (Killick-Kendrick, R. and Peters.
W., eds.), Vol. l, pp. I 120. Academic Press, London.
27 Smith, G. (1973) Unequal crossover in the evolution of
multigene families. Cold Spring Harbor Syrup. Quant.
Biol. 38, 507 513.
28 Bouvier, J., Etges, R. and Bordier, C. (1987) Identification of the promastigote surface proteasc in seven
species of Leishman&. Mol. Biochem. Parasitol. 24, 73
79.
29 Russell, D.G., Ip, H.S. and Medina-Acosta, E. 11991)
Biology of the Leishmania surface protease, gp63. In:
Biology of Parasitism. (Wang, C.C., cd.). pp. 73 85.
Am. Assoc. Adv. Sci.. Washington, DC.
30 McMahon-Pratt, D., Modi, G. and Tesh, ll.B. (1983)
Detection of promastigote stage-specific antigens on
Leishmania mexicana amazonensis developing in the
midgut of Lutzomvia Ion~,ipalpis. Am. J. Trop. Med.
Hyg. 32, 1268 1271.
31 Killick-Kcndrick. R. (1979) Biology of LeishmaHia in
phlebotomine sandflies. In: Biology of the Kinetoplastida. (Lulnsden, W.H.R. and Ewms, D.A., eds.). Vol. 2.
pp. 396 460. Academic Press, London.
32 Adler. S. and Theodor, O. (1929) Observations on
Leishmania ceramodactvli n. sp. Trans. R. Soc. Yrop.
Med. Hyg. 22, 343 356.
33 Adler. S. and Theodor, O. (1930) The behavior of insect
flagellates and leishmanias. Ann. Trop. Med. Parasitol.
24, 193 196.
34 Coburn, C.M., Otteman, K., McNeely. T.. Turco. S.
and Beverley, S.M. (1991) Stable transfection of a wide
range o1" trypanosomatids. Mol. Biochem. Parasitol. 46,
169 179.