* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download The world of proteases Diversity and function
Biochemistry wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Magnesium transporter wikipedia , lookup
Interactome wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Metalloprotein wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Western blot wikipedia , lookup
Protein purification wikipedia , lookup
Specialized pro-resolving mediators wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Proteases in angiogenesis wikipedia , lookup
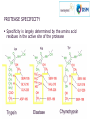
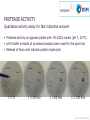
The world of proteases Diversity and function Amsterdam, May 8th 2014 V Glitsø PhD, Senior Department Manager K Pontoppidan PhD, Science Manager Feed Applications Novozymes R&D AGENDA • The world of proteases • Proteases in animal feed 1 The world of proteases DEFINITION OF A PROTEASE Protease = Peptidase = Proteinase = Proteolytic enzyme An enzyme that degrades protein by hydrolysis of peptide bonds Protease OCCURRENCE AND FUNCTIONALITY Analysis of complete genomes has shown that about 2% of proteins in all kinds of organisms are proteases Proteases have many different functions: processing of proteins, protein turnover, cell division, metabolism, toxins… PROTEASE SUBSTRATES Protein is a highly diverse substrate 20 different amino acids to coose from at each position Various types of protein: His Plant proteins Egg white Milk Muscle Hormones Enzymes Crystal structure of porcine trypsin Gly Pro Ala PROTEASE SPECIFICITY Specificity is largely determined by the amino acid residues in the active site of the protease PROTEASE SPECIFICITY Different proteases result in different hydrolysis products Very specific protease Less specific protease Trypsin specific for Arginine and Lysine RONOZYME® ProAct preference for hydrophobic amino acids Hydrolysis is limited (few cuts) Hydrolysis is aggresive (many cuts) PROTEASE CLASSIFICATION Proteases are classified according to: Functionality (reaction catalyzed) Molecular structure and sequence homology (MEROPS) FUNCTIONAL CLASSIFICATION Endo His Gly Pro Ala Exo MEROPS CLASSIFICATION Proteases are divided into 7 major protein families and 196 subfamilies based on molecular structure and sequence homology Catalytic Type (Major protein families) MEROPS families Serine 45 Cysteine Metallo Aspartic Serine residue involved in the catalytic action in the active site 65 59 14 Glutamic 2 Threonine 4 Unknown 7 Total 196 MEROPS Release 9.10 (http://merops.sanger.ac.uk) MICROBIAL PROTEASE DIVERSITY 370,000 SEQUENCES Family A1 - Pepsin Family M14 - Carboxypeptidase A and B Family S1 - Trypsin - Chymotrypsin - Elastase - RONOZYME® ProAct PROTEASE ACTIVITY Protease activity can be measured in many ways using different substrates and different reaction conditions There is not one correct way to measure protease activity An activity number is always dependent on the exact assay conditions Therefore, activity units/protease assays cannot be used to evaluate protease performance – this should be done under real application conditions PROTEASE ACTIVITY Protease activity assays are useful to: Control that our product always contains the same amount of protease activity (QA/QC) As a tool in the development process of proteases Compare the relative (e.g. residual) activity of proteases (e.g. the stability following a challenge such as pelleting or low pH) 2 Proteases in animal feed KEY CHARACTERISTICS FOR A FEED PROTEASE Protease activity Stability in the gut and during processing (pelleting) Synergy with endogenous proteases Compatibility with other feed enzymes PROTEASE ACTIVITY SDS-page analysis to evaluate protease purity and identification Load protein samples here Solution with protein kD Heat + SDS 50 big proteins 35 Visualization e.g. by staining 30 25 small proteins 15 10 PROTEASE ACTIVITY SDS-page analysis to evaluate protease purity and identification kD kD kD 50 50 35 30 25 15 Nocardiopsis Serine protease (>95% pure) 50 35 35 30 30 25 25 15 15 10 10 10 Bacillus Subtilisin proteases >90% estimated to be wheat protein Identities confirmed by proteomics analysis (LC-MS/MS) HOW TO EVALUATE IF A PRODUCT HAS ACTIVE PROTEASE PROTEASE ACTIVITY Quantitive activity assay Relative protease activity (Protease A=1) 2500 2317 Dosed on equal weight (g product/ml) Adjusted for 'in feed' recommendations 2000 1500 Assay conditions adjusted according to protocol for Product A 1448 1000 770 726 500 193 1 0 Ronozyme ProAct 1 Product A Product B 105 168 Product C 182 Product D Casein-FITC substrate, pH 8.3, 15 min, 37°C, very sensitive assay ELN-13-HALL-0016 QUANTITATIVE ACTIVITY ASSAY Protease activity determined using a highly sensitive assay (Casein-FITC substrate, pH 8.3, 15 min, 37°C) Relative protease activity (Protease A=1) 2500 Dosed on equal weight (g product/ml) 2317 Adjusted for 'in feed' recommendations 2000 1500 Assay conditions adjusted according to 3x protocol for Product A 1448 1000 7.5x 770 726 500 193 1 0 Ronozyme ProAct 1 Product Product A A Product B B Product 105 168 Products C Product C 182 Product DD Product ELN-13-HALL-0016 PROTEASE ACTIVITY Qualitative activity assay for fast indicative answers Protease activity on agarose plates with 1% AZCL-casein (pH 7, 22°C) pH 5 buffer extracts of protease products were used for the spot test Release of blue color indicate protein hydrolysis t=0 t = 30 min t = 60 min t = 120 min ELN-14-BERA-0003 PROTEASE ACTIVITY Qualitative assay for activity and stability Skimmed milk plates for a simple test of activity and acid stability Acid instability Acid stability A ProAct A C B C B Dilution in pH 7 buffer Spot on plate Extraction of solid products in pH 6 buffer (1 hour, 100 rpm, 26°C) recovery of liquid fraction Incubation ½ h at pH 3 dilution in pH 7 buffer Incubation of plate (2½ hour, pH 6, 37°C). All extracts diluted 25x in total. Of the tested products RONOZYME® ProAct showed the highest protease activity and it was the only product that was stable at pH 3 (30 min) ELN-13-KPON-0002 SYNERGY WITH ENDOGENOUS PROTEASES In vitro digestion model – a useful tool when screening for complimentary effects Test enzyme Analyse soluble Feed protein Analyse degree of hydrolysis (DH) The results reflect at the same time survival and action in the digestion model SYNERGY WITH ENDOGENOUS PROTEASES Products A, B, C and D represent products claiming protease activity as main activity or as side activity ELN-09-HALL-0008 & ELN-10-LNBR-0065 Improvement relative to control (%) In vitro conditions: - SBM-maize (30:70) - 40°C - pH 3/pepsin: 1hour - pH 7/pancreatin: 4 hours - Protease: 10 x rec. dosage 120 Improvement relative to control (%) Protein solubilization 110 * 115 * * 110 105 100 95 90 85 80 105 * * Degree of hydrolysis 100 95 90 85 * Significant increase (P<0.05, all-pairwise Tukey-kramer HSD) SYNERGY WITH ENDOGENOUS PROTEASES Effect of ProAct on protein sol. of different raw materials in vitro Control 100 100 mg EP/kg Sol protein of total (%) 90 80 70 60 50 40 SBM, Brazil Full fat SBM, Sorghum, Maize, Brazil SBM Full fat Sorghum Maize 1 Brazil Brazil SBM MLRA080001 & HALL100003 MBM, US MBM Feather Wheat Midds Corn DDGS Feather Wheat Corn meal, Brazil (Dakota meal midds DDGS Gold) SYNERGY WITH PANCREATIC PROTEASES Substrate: Commercially toasted SBM Incubation: 3 hours, pH 7, 40°C Enzymes: Pancreatic Trypsin Novo (PTN), RONOZYME® ProAct Analysis: Colorimetric analysis of cleaved peptide bonds with OPA reagent Using ProAct, same effect is obtained with ~½ amount of PTN ELN-13-HALL-0001 COMPATIBILITY WITH OTHER FEED ENZYMES In vitro incubations with enzyme combinations Phytate degradation by phytase and phytase + ProAct (20x) Corn-sbm diet: pH 6/30 min -> pH 3/pepsin/10 min Phytase @ recom. dose, ProAct @ 20x rec. dose Xylan solubilisation by xylanase and xylanase + ProAct (10x) Wheat bran: pH 7/3 hours Xylanase @ recom. dose, ProAct @ 10x rec. dose ELN-11-KPON-0001 & ELN-11-CAAO-0005 CONCLUDING REMARKS Large protease diversity exists: • A protease is not just a protease • Also there are large differences between the products claiming protease activity For feed protease it is important to ensure that the key characteristics are in place: • Detectable protease activity – ability to hydrolyse protein • Stability in gut and during processing • Synergy with endogenous enzymes • Compatibility with other feed enzymes Thank you for your attention MICROBIAL PROTEASE DIVERSITY partate As Serine 4,169 reonine Th 132,946 Metallo ate m uta 674 10,833 ss ncla ifie 140,155 d U 8,778 Gl Peptidases divided on basis of catalytic mechanism Cysteine 59,135 Major protease families bubble areas are proportional to sequence counts 370.000 in total