* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Laboratory #4: Segregation of Traits According to Mendel
Polymorphism (biology) wikipedia , lookup
Behavioural genetics wikipedia , lookup
X-inactivation wikipedia , lookup
Heritability of IQ wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression programming wikipedia , lookup
Genetically modified organism containment and escape wikipedia , lookup
Public health genomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genome (book) wikipedia , lookup
Population genetics wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genetic drift wikipedia , lookup
History of genetic engineering wikipedia , lookup
Designer baby wikipedia , lookup
Genetically modified crops wikipedia , lookup
Microevolution wikipedia , lookup
Quantitative trait locus wikipedia , lookup
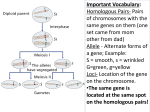
Laboratory #4: Segregation of Traits According to Mendel Background Gregor Mendel was a monk who studied genetic inheritance using pea plants. Peas are self-pollinating plants, so after multiple generations (without interference) of self-fertilization they become homozygous for all their important qualitative genes and are known as a pure line. Mendel crossfertilized different homozygous pea lines to see what would happen. As a result of his work with peas, Mendel concluded that physical traits are passed from one generation to the next as discrete units, which we now call genes. Genes are a section of DNA that codes for a protein. These proteins create particular traits, often through complicated interactions and pathways. Alleles are possible forms a gene can take. In peas as in humans and other diploid species, there are two alleles for each gene. These alleles may be the same (homozygous) or they may be different (heterozygous). Alleles can be either dominant or recessive. An allele that is dominant will be expressed any time it is present in either the homozygous dominant or heterozygous state. A recessive allele will only be expressed if no dominant alleles are present and the recessive allele is in the homozygous recessive state. The physical appearance of an organism is its phenotype (wrinkled, smooth, yellow, green). This appearance is coded for by the alleles present for the gene. The actual alleles and genes that determine the phenotype are the genotype of a trait, and are illustrated using capital and lower case letters. Let us consider Mendel’s round pea x wrinkled pea example. In the first generation (F1), all the peas resulting from a cross between wrinkled and round, were round. Thus the round pea allele (gene) is dominant to the wrinkled pea allele. Taking a look at the F2 generation of progeny in the smooth versus wrinkled pea example, we can see that 75% of the progeny are smooth, which also demonstrates that the round characteristic is dominant over the wrinkled. We can illustrate this by assigning the capital letter ‘S’ to the dominant allele controlling the round pea and the small letter ‘s’ to the recessive allele that results in the wrinkled trait. Mendel’s experiment with smooth and wrinkled peas would look like: Crossing the parents to create the F1 generation. Homozygous smooth ‘S’ peas crossed to homozygous wrinkled ‘s’ peas SS x ss s S Ss S Ss s Ss Ss 100% of the F1 generation have the heterozygous genotype ‘Ss’ and will have the smooth phenotype as a single copy of the ‘S’ allele is sufficient to attain round peas. Self-pollinating the F1 generation plants to create the F2 generation. Heterozygous smooth ‘Ss’ by heterozygous smooth ‘Ss’ Ss x Ss S S SS s Ss S Ss Ss From this we have that 25% of the progeny have the ‘SS’ genotype and are hence round (homozygous), 50% have the ‘Ss’ genotype and are also round (heterozygous), and 25% of the progeny have the ‘ss’ genotype and are hence wrinkled. Pulling like phenotypes together we have that 75% (25% + 50%) of the F2 generation will have the smooth phenotype (genotype ‘Ss’ and ‘SS’) and 25% of the F2 generation will have the wrinkled phenotype (homozygous recessive genotype ‘ss’). A number of agronomically important characteristics (i.e. yield, protein content, drought resistance, etc) are not controlled by a single gene but are controlled by many, sometimes thousands. Rather than produce discrete classes (i.e. round v wrinkled) variation within progeny for quantitatively (many genes) inherited characteristics show continuous variation. To relate this concept to Mendelian genetics, if the smooth versus wrinkled characteristic of Mendel’s peas was continuous and therefore controlled by a number of genes, Mendel would have observed phenotypes intermediate to either of the parents (i.e. a little bit wrinkled, or almost round) when he crossed smooth and wrinkled pure lines. This experiment will demonstrate Mendelian segregation using an easy to observe trait (Roundup herbicide resistance). The species that will be used is canola (Brassica napus L.). One of the parents used in this crossing study had been transformed to have a gene that confers resistance to glyphosate, the active ingredient in Roundup herbicide. This cultivar (genotype) was crossed to a second genotype which did not have resistance to Roundup herbicide. The crossing and F1 part of the study was completed last year. To complete this genetic study you will be provided with F2 seed from the cross between a glyphosate (Roundup) resistant parent and glyphosate susceptible parent. You will use the progeny from these seeds to determine whether the inheritance of Roundup resistance is controlled by a single gene according to Mendel’s Laws of Segregation. Herbicide susceptibility will be tested by spraying the plants with glyphosate (Roundup). Objectives Observe segregation in Roundup resistant canola. Determine whether glyphosate resistance is controlled by a single gene. Materials and Methods Each group will plant 80 F2 seeds of unknown genotype along with 10 seeds from the susceptible parent and 10 seeds from the resistant parent in a seedling flat. Three weeks after planting, when plants are at approximately at the three to four leaf stage, each group will spray their plants with glyphosate solution. In the following weeks the resistant and susceptible plants will be noted. In two weeks, when herbicide damage symptoms are clearly visible, results will be compiled as a class. Calulating Chi-square It is often necessary to test whether the distribution of progeny we observe from a cross does indeed follow the frequency expected given a simple Mendelian inheritance. This can be achieved by doing an appropriate Chi-square calculation. Chi-square calculations compare the expected phenotypic or genotypic ratio to the observed results of the experiment to mathematically determine the distance between an acceptable fit for the ratio to the results and the actual fit of the ratio. The formula to calculate Chi-Square is: X2obs=Σ[((obss-exps) 2/exps)+((obsw-expw) 2/expw)] Where obss = Observed number of smooth peas, exps = Expected number of smooth peas, obsw = Observed number of wrinkled peas, expw = Expected number of wrinkled peas. Expected values are calculated by multiplying the total number of peas by the expected percentage of the peas that should have the trait (i.e. 75% should be smooth). X2table is then looked up on a Chi-Square chart. These can be found in any statistics book, and will be provided for you in a lab handout. When looking up a chi-square value, you must know the amount of certainty you want and the degrees of freedom. Scientific experiments are typically judged at the 95% certainty level. The degrees of freedom is the number of independent categories (usually the number of categories minus one). In Mendel’s pea experiment there are two categories (smooth and wrinkled) so the degrees of freedom would be one, making the X2 value 3.84. If the ratio fits the data (in this experiment, if there is a 3:1 ratio of smooth versus wrinkled peas) then, X2obs < X2table, and the ratio is said to have a good fit. This means the results are as expected based on the laws of Mendelian genetics. If the ratio is not a good fit for the data then, X2obs > X2table, the data is said to not fit the 3:1 ratio. This means that the results of the experiment are not as expected based on the laws of Mendelian genetics. Assignment 1. What is the difference between a homozygous genotype and a heterozygous genotype? 2. Why would you want homozygous alleles in crops? 3. What was your group's ratio of resistant:susceptible? 4. What was the class' ratio of resistant:susceptible? 5. What are the results of a chi-square test of the resulting ratio? 6. Based on your results is glyphosate resistance a single-gene trait? Why? 7. What did you learn in this laboratory? 8. How would you change this laboratory?