* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Classification of plant-pathogenic mycoplasma
Genetically modified organism containment and escape wikipedia , lookup
Bioinformatics wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
DNA vaccination wikipedia , lookup
Social sequence analysis wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Genetic engineering wikipedia , lookup
Designer baby wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
SNP genotyping wikipedia , lookup
Molecular cloning wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Genomic library wikipedia , lookup
Multilocus sequence typing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene prediction wikipedia , lookup
Metagenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Journal of General Microbiology (1993), 139, 519-527.
Printed in Great Britain
519
Classification of plant-pathogenic mycoplasma-like organisms using
restriction-site analysis of PCR-amplified 16s rDNA
BERNDSCHNEIDER,
ULRICHAHRENS,~
BRUCEC. KIRK PAT RICK^ and ERICHSEEMULLER'
*
'Biologische Bundesanstalt, Institut fur PJlanzenschutz im Obstbau, 0-6915 Dossenheim, Germany
2Department of Plant Pathology, University of California, Davis, C A 95616, USA
(Received 3 June 1992; revised 26 October 1992; accepted 5 November 1992)
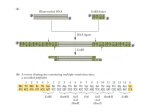
A method has been developed to amplify the 16s rRNA gene of plant-pathogenic mycoplasma-like organisms
(MLOs) from infected plant material using the polymerase chain reaction (PCR). The procedure is dependent on
the presence of a BcZI restriction site in the 16s rDNA of chloroplasts but not in that of the MLOs. This difference
permits the specific amplification of the 16s rDNA of the MLOs from BcZI-digested total DNA from infected
plants using primers from conserved regions of this gene. In this study 16s rDNA was obtained from 52 MLO
isolates from herbaceous dicots and monocots as well as woody plants. Digestion of the 16s rRNA genes using A M
endonuclease revealed seven restriction patterns, which were used to group the isolates examined. Group I, which
is also characterized by the presence of two KpnI sites, consisted of 31 isolates, most of which are from herbaceous
dicots. Isolates assigned to groups I1 to VI were mostly from woody plants, while the isolates of group VII were
from monocots or obtained from a leafhopper. The restriction patterns varied little within groups; however, four
group I isolates and one group IV isolate differed slightly from the typical patterns of these groups as a result of
a deletion or a slight shift of one restriction site. The groupings uncovered by Ah1 restriction were also obtained
by digesting the 16s rDNA with RsaI endonuclease. However, some atypical patterns were observed within group
V isolates. The groups described on the basis of restriction digest data were supported by sequence analysis. With
one exception, the 16s rDNA of isolates within the same group exhibited 97.8 to 99-5YOhomology while those of
different groups showed 89.6 to 92.0% homology.
Introduction
Mycoplasma-like organisms (MLOs) are nonculturable,
parasitic prQkaryotes of the class Mollicutes associated
with diseases of several hundred plant species (McCoy et
al., 1989). Until recently, differentiation and characterization was mainly based on host range and the
symptoms induced in natural hosts and in the experimental host Catharanthus roseus (L.) G . Don (periwinkle) (Marwitz, 1990). However, with the introduction
of serological and nucleic acid hybridization methods
into plant mycoplasmology, more reliable and specific
means are available to characterize MLOs. The development of techniques to obtain MLO DNA from
infected plants and insect vectors and the cloning of
MLO DNA have greatly enhanced this work. A number
*Author for correspondence. Tel. 49 6211 85238; fax 49 6221
86 1222.
Abbreviation : MLO, mycoplasma-like organism.
0001-7614 0 1993 SGM
of recent papers on dot and Southern hybridization has
contributed to our better understanding of the relatedness
of the MLOs (Bertaccini et al., 1990; Bonnet et al., 1990;
Lee & Davis, 1988; Lee et al., 1990; Kuske et al., 1991).
Based on Southern hybridization with a DNA fragment
of an MLO associated with aster yellows, a differentiation between organisms inducing decline symptoms and
those causing floral virescence has been proposed (Kuske
et al., 1991). Although several organisms have been
differentiated using these methods they are limited by the
fact that undefined DNA fragments have been used as
probes.
In contrast to undefined genomic DNA fragments, the
16s rRNA gene is a universal character which provides
valuable molecular information on MLOs. This gene
shows regions which are highly conserved among the
prokaryotes while other regions show considerable
variation, thus permitting phylogenetic and taxonomic
studies (Stackebrandt, 1991). Recently, 16s rRNA sequences have been used for the phylogenetic analysis and
classification of culturable mollicutes (Weisburg et al.,
1989) and to elucidate the phylogeny of two MLOs (Lim
Downloaded from www.microbiologyresearch.org by
IP: 88.99.165.207
On: Fri, 16 Jun 2017 11:38:44
520
B. Schneider and others
Table 1. Origin and source of the MLO isolates and the symptoms they induce in
periwinkle
MLO
code
AAY
ACLR
AKV
AP
ASHY
AT
AV2 192
AV2226
AYW
BGWL
BVK
COL
CVA
CVB
CVL
CVT
DEV
DIV
EAY
EY
FDI
GVX
HYDP
KV
KVE
MOL
OAY
PARM
PER
PLN-V6
PPER
PRIVA
PRIVB
PRIVC
PSER
PVM
PVW
PYLR
RCAE
RV
SAFP
SAS
SAY
SBB
SCWL
STOL
STOLF
SUNHP
TBB
ULW
VAC
wx
Origin
American aster yellows
Apricot chlorotic leaf roll
Virescence of Aquilegia alpina L.
Apple proliferation
Ash yellows
Apple proliferation
Aster yellows
Aster yellows
Eastern American aster yellows
Bermudagrass white leaf
Leafhopper-borne (Psammotettix cephalotes)
Latent in Cuscuta odorata Ruiz et Pav.
Leafhopper-borne (species not determined)
Leafhopper-borne (species not determined)
Catharanthus virescence
Catharanthus virescence
Virescence of a Delphinium hybrid
Virescence of Diplotaxis erucoides (L.) DC
Aster yellows
Elm yellows
Flavescence dorhe
Green Valley strain of X-disease
Hydrangea phyllody
Clover phyllody
Clover phyllody
Molikre’s disease of cherry
Virescence of Oenothera hookeri
Apricot decline
Peach decline
Plum leptonecrosis
Peach decline
Virescence of primrose (Primula sp.)
Virescence of primrose (Primula sp.)
Virescence of primrose (Primula sp.)
Decline of Prunus serrulata Lindl.
Virescence of Plantago coronopus L.
Virescence of Plantago major L.
Peach yellow leaf roll
Rubus stunt of R. caesius L.
Rape virescence
Safflower phyllody
Sandal spike
American western aster yellows
Big bud of Solanum marginatum L.
Sugarcane white leaf
Stolbur of Capsicum annuum L.
Stolbur of Lycopersicon esculentum Mill.
Sunhemp phyllody
Tomato big bud
Witches’ broom of Ulmus carpinifolia Gled.
Witches’ broom of Vaccinium myrtillus L.
Western X-disease
Country/
state
Florida
Spain
Germany
Italy
New York
Germany
Germany
Germany
New Jersey
Thailand
Germany
?
Germany
Germany
Peru
Thailand
Germany
Spain
Germany
New York
Italy
California
Belgium
Germany
England
France
USA
Germany
Italy
Italy
Germany
Germany
Germany
Germany
Germany
Germany
Germany
California
Germany
France
Israel
India
California
Ecuador
Thailand
Croatia
France
Thailand
Australia
France
Germany
California
Source*
1
2
3
4a
5
3a
3
3
6
7
8
8a
8
8
9
7
3b
10
3
5a
4
11
12
3
11
13
14
7
15
4
7
3c
3c
3c
7
3
8
11
7
16
17
18
19
3d
7
20
21
7
11
22
3e
23
Symptom
group t
A
A
A
D
D
D
B
B
B
D
D
A
A
A
A
C
B
B
D
A
D
A
A
A
A
A
A
C
D
A
B
C
D
B
A
A
A
A
A
A
A
A
D
D
* Collected and/or transmitted to periwinkle or Coleus blumei, or provided by: 1, R. E. McCoy, University of
Florida, Fort Lauderdale, USA (via 3); 2, G. Llacer, IVIA, Moncada-Valencia, Spain (via 13); 3, R. Marwitz,
Biologische Bundesanstalt, Berlin, Germany (3a Marwitz et al., 1974, 3b Marwitz & Petzold, 1976, 3c Marwitz
& Petzold, 1983, 3d Marwitz et al., 1979, 3e Marwitz et al., 1987); 4, L. Carraro, Universita degli Studi, Udine,
Italy (4a Carraro et al., 1988); 5, W. A. Sinclair, Cornell University, Ithaca NY, USA (5a Sinclair et al., 1976);
6, R. F. Whitcomb, USDA-ARS, Beltsville MD, USA (via 11); 7, collected by the authors; 8, W. Heintz,
Biologische Bundesanstalt, Dossenheim, Germany (8a Heintz, 1989); 9, C. E. Fribourg, International Potato
Center, Lima, Peru (via 3); 10, P. Moreno, IVIA, Moncada-Valencia, Spain (Moreno et al., 1985) (via 3); 11,
M. F. Clark, Horticulture Research International, East Malling, UK; 12, W. Welvaert, Rijksuniversiteit, Gent,
Belgium (Welvaert et al., 1975) (via 3); 13, F. Dosba, INRA, Bordeaux, France; 14, B. B. Sears, MSU, East
Lansing, USA (Sears & Klomparens, 1989); 15, A. Ragozzino, Universita di Napoli-Portici, Naples, Italy (via 13);
Downloaded from www.microbiologyresearch.org by
IP: 88.99.165.207
On: Fri, 16 Jun 2017 11:38:44
MLO classiJication
& Sears, 1989; Kuske & Kirkpatrick, 1992). Both
sequence and restriction enzyme analysis of 16s rDNA
have been used in taxonomic studies on mollicutes
(Gobel et al., 1987; Laigret et al., 1990; Taschke et al.,
1990) and other prokaryotes (Grimont & Grimont,
1986; Bouvet et al., 1991;Gurtler et al., 1991).The study
of the 16s rRNA gene is greatly facilitated by the
application of polymerase chain reaction (PCR) technology using primers that allow amplification of prokaryote 16s rDNA. Here we report a method which
amplifies the 16SrRNA gene from MLOs and can be
used to group them on the basis of restriction enzyme
analysis of 16s rDNA.
Methods
Sources of MLOs and MLO 16SrDNA sequences. Sources of the
MLOs examined and the codes used to describe them are listed in Table
1. Isolate EAY was maintained in the greenhouse in Coleus blumei
Benth. by cuttings. PARM, PPER, PSER and RCAE were obtained
from diseased apricot, peach, flowering cherry (Prunus serrulata Lindl.)
and Rubus caesius L., respectively, grown in the experimental field of
the Dossenheim institute. BGWL and SCWL were obtained from
diseased bermudagrass and sugarcane, respectively, collected near
Bangkok, Thailand. All other isolates were maintained in an insectproof greenhouse in periwinkle by graft transmission. With the
exception of the naturally infected isolates, CVL and CVT, the
periwinkle-maintained MLOs were originally transmitted to this host
with Cuscuta spp. bridges or by leafhoppers. The symptoms induced in
periwinkle are indicated in Table 1. 16s rDNA sequences of the
Oenothera (OAY-) (Lim & Sears, 1989) and western aster yellows
(SAY-) MLOs (Kuske & Kirkpatrick, 1992) were included in the study
for comparison.
Other prokaryotes. Isolates of Agrobacterium tumefaciens (strain At
l), Claoibactermichiganensis (strain C 2 140), Erwinia amylooora (strain
Ea. 6/6), and Xanthomonas campestris (strain Xc 314) (all obtained
from W. Zeller, Biologische Bundesanstalt, Dossenheim) were grown
on nutrient glycerol agar slants. Escherichia coli (strain XLl blue,
Stratagene) and Spiroplasma cirri (strain R8A2, obtained from C.
Saillard, INRA Bordeaux, France) were cultivated in LB medium
(Maniatis at al., 1982) and BSR medium (Bovk & Saillard, 1979),
respectively. Overnight growth from the cultures on solid media
suspended in water and similar-aged liquid cultures of E. coli and S.
cirri were used without further treatment for in vitro amplification of
16s rDNA as described below.
PCR amplification. Five different primers from conserved regions of
the 16s rRNA gene were used. The pair fDl and rP1 (Weisburg et al.,
52 1
1991) primed proximal to the 5' and 3' termini, allowing the
amplification of nearly the entire 16s rRNA gene. The three internal
primers consisted of the forward primer fA extending from position 759
to 778 of the OAY-MLO (Lim & Sears, 1989), the reverse primer rA
extending from position 1316 to 1297 of the same organism (Ahrens &
Seemuller, 1992), and the reverse primer rC which is complementary to
primer fA.
DNA from healthy and diseased plants was obtained by using an
MLO enrichment procedure as described previously (Ahrens &
Seemiiller, 1992). Five microlitres of such DNA preparations were
digested with 5 U of BclI restriction endonuclease (Amersham) in a
total volume of 20p1. Five microlitres of the digest or of bacterial
suspension were used to amplify DNA in a 50 p1 reaction containing
125 p~ of the four dNTPs, 0.5 p~ of each of the primers, and 1 U of
Taq polymerase (Boehringer-Mannheim). PCR conditions consisted of
30 cycles of 30 s at 95 "C, 30 s at 50 "C, and 60 s at 72 "C, plus one
additional cycle with a 4 min chain elongation. After amplification,
5 pl of the product was digested with BclI as described above and was
then separated by electrophoresis in a horizontal 1 % (w/v) agarose gel
in TAE buffer (40 mM-Tris/acetate, 1 mM-EDTA, pH 8.0). From the
band containing the desired 16s rDNA some material was removed
from the gel with a hypodermic syringe and was, without further
purification, amplified again as described above. The purity of the final
product was examined by another BclI digestion followed by agarose
gel electrophoresis. 16s rDNA of healthy plants was amplified without
BclI digestion. All amplifications were performed using a Thermocycler
60 (bio-med).
Restriction digest and gel electrophoresis. The final amplification
products were digested with AluI, RsaI, EcoRI or KpnI, following the
manufacturer's instructions (Amersham). Five microlitres of Ah1 and
RsaI digests were resolved on vertical 5 or 8 % (w/v) polyacrylamide
gels in TBE buffer (89 w-Tris/borate, 89 mM-boric acid, 2 mMEDTA, pH 8.0). The EcoRI and KpnI digests were separated by
electrophoresis in 1% (w/v) agarose as previously described. The DNA
was visualized under UV light after staining with ethidium bromide.
DNA sequencing. PCR-amplified 16s rDNAs from the AAY-,
ACLR-, ASHY-, AT-, EY-, PPER- and ULW-MLOs were cloned in
Bluescript M 13+ (Stratagene) using standard procedures; ligation was
performed according to Marchuk et al. (1991). One strand of the cloned
16s rDNA was sequenced with the Sequenase kit (US Biochemical)
following the manufacturer's instructions. Two universal primers
priming near the multiple cloning site (T3 and SK, Stratagene), one
internal primer designed by Lane et al. (1985) extending from position
532 to 515 of the 16s rRNA gene of E. coli, another reverse primer
designed by F. Laigret (personal communication, position 1068 to 1049
of E. coli), and the two internal reverse primers rA and rC described
previously were used.
Analysis of data. Sequences of the 16s rRNA genes of the following
organisms were compared by multiple alignment using the Clustal
program (Higgins & Sharp, 1988): OAY- (EMBL/GenBank accession
15, G. Marchoux, INRA, Avignon-Montfavet, France (Marchoux & Giannotti, 1971) (via 3); 17, M. Klein,
Volcani Center, Bed Dagan, Israel (Klein, 1970) (via 3); 18, J. Dijkstra, Agricultural University, Wageningen,
Netherlands (Dijkstra & Lee, 1972) (via 3); 19, B. C. Kirkpatrick, University of California, Davis, USA (Kuske
& Kirkpatrick, 1992); 20, D. Sutic, University of Zagreb, Croatia (via 3); 21, M.-T. Cousin, INRA, Versailles,
France; 22, G. Morvan, INRA, Avignon-Montfavet, France (via 13); 23, D. D. Jensen, University of California,
Berkeley (Jensen, 1986).
t According to the predominant symptoms in periwinkle : A, virescence, phyllody (typical for clover phyllody
and stolbur) ; B, virescence, phyllody, elongated and etiolated internodes (typical aster yellows symptoms) ; C,
small and faintly coloured flowers, elongated and etiolated internodes (atypical aster yellows symptoms) ; D,
reduced flower size, leaf and flower malformations, no virescence, phyllody or elongated and etiolated internodes
('decline MLOs').
Downloaded from www.microbiologyresearch.org by
IP: 88.99.165.207
On: Fri, 16 Jun 2017 11:38:44
522
B. Schneider and others
no. M30970), WX- (L04682), SAY- (M86340), AAY- (X68373),
ACLR- (X68338), ASHY- (X68339), AT- (X68375), EY- (X68377),
PPER- 668374) and ULW- (X68376) MLOs, and S. citri (M23942).
Analysis for the presence of BclI, AluI and RsaI restriction sites was
performed with 16s rDNA of Spiroplasma apis (M23937), Mycoplasma
hominis (M2447), M . hyopneumoniae (Y00149), M . capricolum
(X00921), Acholeplasma laidlawii (M23932), Anaeroplasma abactoclasticum (M25050), E. coli (V00348), and Clostridium innocuum
(M23732). The 16s rDNA of the chloroplasts of Glycine max (X06428),
Nicotiana tabacum (V00165), Pisum sativum (M30826), Spinacea
oleracea (101440) and Sinapis a h a (X04182) were also included in
restriction site analysis. All sequences of the culturable prokaryotes and
the chloroplasts, as well as those of the OAY- and SAY-MLOs, were
available in the EMBL Data Library, Heidelberg, Germany.
Results
baceous dicots or were obtained from leafhoppers (Table
1). All isolates of group I showed five restriction sites at
positions a, d, e, f and g (Fig. 1). Group I contains isolate
MOL and the stolbur-type isolates STOL, STOLF and
TBB, which differ from the other group members by a
g h
I1 (ACLR)
Amplijication of 16s rDNA
Sequence comparisons with the 16s rRNA genes of the
OAY-, WX-, AT- and AAY-MLOs and the chloroplasts
examined revealed the presence of a BclI restriction site
in the chloroplasts but not in the gene of the MLOs. To
obtain a specific PCR amplification of the 16s rDNA of
the MLOs, the DNA from infected plants was digested
with BclI before amplification. As the digest was usually
incomplete, the amplification products were also BclIrestricted. Gel electrophoresis resolved one fragment
approximately 1500 bp in size representing the 16s
rRNA gene of the MLOs. The cleaved 16s rDNA of the
chloroplasts appeared as two fragments approximately
800 and 700 bp in length.
i i i i - h u
111 (ASHY)
ij
f
l
IV(EY)
V(AT)
VI(wx)
VII (SCWL)
1
Restriction and sequence analysis
Fig. 1. A h 1 restriction map of 16s rDNA depicting the seven (I-VII)
different restriction profiles that occur among the MLOs examined.
Representative isolates of the seven groups are given in parentheses (see
Table 1 for MLO code). The figures given in group I correspond to the
sequence positions of the OAY-MLO.
The 52 isolates examined were grouped according to the
presence of AluI and RsaI restriction sites. All sites of
groups I to VI of which complete (AAY, ACLR, AT,
OAY, PPER, SAY, WX) or partial (ASHY, EY, ULW)
sequences of representative isolates were available could
be determined. With the exception of the RsaI sites
yielding very small fragments, all sites of group VII were
also determined using internal primers. The sizes of the
amplified rDNA sequences and of the restriction fragments differed slightly due to small deletions or insertions. However, the position of the restriction sites could
be determined by aligning the sequences with the
analogous sequence of the OAY-MLO to which all MLO
positions given in this paper correspond.
Restriction digestion of the amplified 16s rDNA with
AluI revealed seven different profiles among the MLO
isolates (Figs 1 and 2), which were used to divide the
isolates into seven major groups (Table 2). Group I is the
largest and includes 3 1 isolates. With the exception of the
periwinkle-maintained isolates HYDP, MOL, SAS and
PER from woody hosts, they all originate from her-
Fig. 2. AluI restriction profiles of 16s rDNA from MLOs representing
six of the seven groups established. AT to PARM, group I; RCAE to
EY, group IV; ACLR and PLN-V6, group I1 ;ASHY, group 111;AAY,
group I; VAC to SUNHP, group VI; C . ros., healthy periwinkle; S .
citri, Spiroplasma citri. See Table 1 for MLO codes.
Downloaded from www.microbiologyresearch.org by
IP: 88.99.165.207
On: Fri, 16 Jun 2017 11:38:44
523
MLO classijication
Table 3 . RsaI restriction maps of PCR-amplijied 16s
rDNA of MLOs representing the seven groups evidenced
by AluI digestion
Groups and representative isolates
Site and
position*
k
425
482
819
843
863
879
883
956
1381
I
I1
I11
IV
AAY ACLRASHY EY
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
-
-
V
AT
(-)
VII
VI
WX SCWL
+
+
+
-
+ +
+
+ - + ?
+ + + ?
+
+
?
+ + +- ++ + + + +
*Corresponding to the OAY-MLO (Lim & Sears, 1989). +, -,
1
m
n
o
p
q
r
s
Fig. 3. AluI restriction profiles of 16s rDNA of MLO isolates of group
I showing that one fragment (arrow) of the isolates TBB to STOLF is
slightly smaller than those of the typical isolates PRIVA to SBB. See
Table 1 for MLO codes.
Table 2. Grouping of the MLO isolates examined based
on restriction analysis of AluI- and RsaI-digested 16s
rDNA
Group members showing atypical
results
Group
I
I1
111
IV
V
VI
VII
Typical isolates*
Ah1 digest
RsaI digest
AAY, AKV, AV2192, MOL, STOL,
AV2226, AYW, COL, STOLF, TBB
CVA, CVB, CVL, CVT,
DEV, DIV, EAY,
HYDP, KV, KVE, OAY,
PER, PRIVA, PRIVB,
PRIVC, PVM, PVW,
RV, SAFP, SAS, SAY,
SBB
ACLR, PLN-V6
ASHY
EY, RCAE, ULW
AP,AT
MOL, STOL,
STOLF, TBB
FDI, GVX, PYLR,
VAC, WX
BGWL, BVK, SCWL
SUNHP
PARM, PPER,
PSERT
SUNHP
*See Table 1 for MLO code.
t AluI restriction profiles of these isolates are identical to that of the
AP- and AT-MLO.
slightly shorter 3’ fragment (Fig. 3). Restriction analysis
of a fragment amplified with the internal primers fA and
rA revealed that these isolates have a deletion of
estimated size 10 bp near restriction site g.
Most isolates of groups I1 to VI were from woody
hosts (Table 1). Group I1 differs from group I by the lack
of AluI restriction site e. The ASHY-MLO, which
+
+
+
+
+
+
-
-
restriction site present or missing, respectively; ?, site could not be
determined because restriction fragments too small; (-) site present in
isolates PARM, PPER, and PSER of Group V.
represents group 111, lacks restriction sites d and f but
shows restriction sites b and i. Groups IV to VII lack
restriction site a. In addition, group IV lacks restriction
sites d and f and is the only group with restriction sites
h and j. Restriction site c occurs in group V only. Group
VI is characterized by the presence of only three AluI
restriction sites, at positions b, f and g (Figs 1 and 2). The
SUNHP-MLO differs, probably due to a downstream
shift of restriction site b, from the other isolates of group
VI by a smaller fragment between sites b and f and a
larger 5’ fragment. Group VII (data not shown in Fig. 2)
includes two MLOs from the monocot hosts sugarcane
and bermudagrass as well as the leafhopper-borne isolate
BVK. With the exception of the missing restriction site c,
this group shows the same pattern as group V. The only
AluI restriction site common to all the MLOs is g (Fig.
1)RsaI restriction analysis recovered identical groupings
to those revealed by AluI digestion (Tables 2 and 3, Fig.
4). With the former enzyme, a total of nine restriction
sites (k to s) were found, of which the three at positions
m, q and s were common to all MLOs examined. The
isolates of group I have eight restriction sites and show
the same profile. As with the patterns obtained after AluI
digestion, isolate MOL and the stolbur-type isolates
STOL, STOLF and TBB differ from the other MLOs of
this group by a deletion near the 3’ terminus, resulting in
a shorter 425 bp-fragment between sites r and s. The
isolates within the groups I1 and IV, respectively, were
homogeneous. Isolate ASHY (group 111) differs from
group IV by the lack of restriction site r. In group V, the
Prunus isolates PARM, PPER and PSER differ from the
apple isolates AP and AT by an additional restriction site
at position k. In group VI, all isolates show the same
Downloaded from www.microbiologyresearch.org by
IP: 88.99.165.207
On: Fri, 16 Jun 2017 11:38:44
524
B. Schneider and others
Fig. 4. RsaI restriction profiles of 16s rDNA from MLOs representing
the seven groups established. AAY and STOL, group I (AAY has two
fragments 425 bp in length while for STOL one of them is slightly
smaller); ACLR, group 11; ASHY, group 111; EY, group IV; AT and
PPER, group V (PPER has an additional restriction site); VAC and
SUNHP, group VI (SUNHP varies in the two uppermost fragments);
SCWL, group VII; C. ms., healthy periwinkle. See Table 1 for MLO
codes.
Table 4. Sequence homology (YO)of the 16s rDNA of
various MLOs
Discussion
See Table 1 for MLO code.
AAY
OAY
SAY
ACLR
AT
PPER
site at either position 488 or 959. In addition, the two
organisms of group I1 have one KpnI site at position 959.
The differences between the groups and the homogeneity within individual groups was also shown by
comparing the homology of the total sequences of the
amplified rRNA genes (Table 4). Thus, isolates of the
same group exhibit 98.4 to 99-5YOhomology while those
of different groups usually differ by about 10%.
However, isolate ACLR of group I1 is highly homologous to the group I MLOs. Isolate PPER, which
produced an atypical RsaI restriction profile, showed a
homology of 97.8% with AT-MLO, another group V
isolate.
Sequence analysis showed that all the walled and wallless prokaryotes examined differ in the A h 1 and RsaI
restriction patterns of the 16s rDNA from those of the
MLOs included in this study. Also, the 16s rRNA genes
of the plant pathogenic bacteria Xanthomonas campestris, Erwinia amylovora, Clavibacter michiganensis
and Agrobacterium tumefaciens showed different profiles
than the MLOs. The RsaI patterns of E. coli and S . citri
are shown in Fig. 4.
In both the AluI and RsaI restriction profiles of a
number of MLOs, a DNA fragment approximately 100
bp in size became evident which is not part of the 16s
rRNA gene (Figs 2 and 4). Its origin remains obscure.
OAY
SAY
ACLR
AT
PPER
WX
98.4
98.7
99.5
97.9
98.4
98.0
90.0
92.0
90.4
90.1
91.4
92.6
91.4
90.9
97.8
89-6
90.7
90.0
90.0
90.6
90-9
profile except SUNHP, which showed slight differences
in the AluI digestion and has a 5’ fragment that is
approximately 15 bp longer than that of the other
isolates while the fragment between position q and s is
shorter to the same degree. SCWL shows the same major
fragments as AT and AP. However, the position of RsaI
sites at the Rsa site aggregation in the middle of the gene
could not be determined with the electrophoresismethods
used.
All isolates showed a unique EcoRI restriction site in
the 16s rDNA at position 669 of the OAY-MLO. Also,
a unique NruI site at position 1340 was detected in all
organisms from which sequencing data were available.
Isolates of group I showed two KpnI sites at positions
488 and 959 which were absent in the other groups. The
only exception was CVB (group I), which has one KpnI
The 16s rRNA gene is the most widely used sequence in
taxonomic studies on the prokaryotes. However, the
study of 16s rRNA sequences in MLOs presents problems as they have not, as yet, been cultured in vitro. With
the method described in this paper, 16s rDNA of the
MLOs can readily be obtained by PCR amplification
without recourse to in vitro cultivation. Examination of
the amplified fragment by restriction and sequence
analysis showed that it represents the authentic 16s
rDNA of the MLOs and not the gene of other plantassociated prokaryotes or chloroplasts. Also, there is
strong evidence that the amplified sequence was from
one organism or at least from organisms of the same
group because the restriction patterns of the isolates
assigned to a group or a subgroup were uniform and
there was no indication of mixed patterns. However, the
presence of a second organism from a different group
cannot be excluded, if it was present in low numbers
relative to the organism that was amplified. The results
obtained do not give information on the number of
rRNA operons. The fact that 16s rDNA of any MLO
would have been amplified with the primers used and
that the restriction patterns were typical for one rRNA
gene indicates that the MLOs examined have either only
one rRNA operon or the copies of the 16s rRNA gene of
Downloaded from www.microbiologyresearch.org by
IP: 88.99.165.207
On: Fri, 16 Jun 2017 11:38:44
MLO classijication
one organism yield similar restriction patterns. The
culturable mollicutes have either one or two rRNA
operons (Razin, 1989), and two occur in the OAY-MLO
(Lim & Sears, 1989).
The MLOs examined originate from four continents
and are associated with diseases of woody plants and of
herbaceous monocots and dicots. In addition, the
organisms induce very different symptoms in periwinkle.
On the basis of the restriction patterns of the 16s rDNA,
the organisms included in this study could be divided
into seven groups. About 60% of the isolates were
assigned to group I, in which MLOs from all four
symptom groups are represented (Table 1). Most of these
MLOs were from herbaceous dicots and include the
agents of aster yellows, clover phyllody, periwinkle
virescence, and stolbur and/or big bud of solanaceous
plants.
There are results from hybridization experiments
which support the interrelatedness of the obviously
diverse MLOs of group I. B. Schneider (unpublished
results) hybridized Southern blots of DNA from most of
the organisms included in this study with chromosomal
DNA probes of the AAY-MLO. Under moderate
stringency conditions these probes hybridized to most of
the MLOs included in group I. Kuske et al. (1991) found
homology between DNA probes from an AY-MLO and
the stolbur isolate STOL as well as several isolates that
induce AY symptoms in periwinkle. Also, probes from
another AY-MLO cross-hybridized with DNA of the
tomato big bud (BB)-MLO and of an MLO causing
virescence in C . roseus (Lee & Davis, 1988). On the other
hand, DNA fragments of the BB-MLO hybridized with
DNA of the MLOs associated with clover phyllody and
a virescence of C . roseus (Lee et al., 1990).
In contrast to group I, the MLOs within the other
groups are more uniform with regard to symptom
induction and host range. The two isolates of group I1
(ACLR and PLN-V6) are considered to be identical or
closely related because they induce similar symptoms in
periwinkle and showed close relationship in Southern
hybridization experiments (Ahrens et al., 1992). The
ASHY- and the EY-MLOs induce similar symptoms in
periwinkle but showed some differences in the restriction
patterns of 16s rDNA and were, for that reason, assigned
to different groups (I11 and IV). This distinction appears
appropriate as Bertaccini et al. (1990) and Davis et al.
(1992) found little cross-hybridization between these two
isolates. Probes from the ASHY- and the EY-MLO did
not cross-hybridize to DNA of the MLOs associated
with AY, BB, and virescence of C . roseus. The ULWMLO, the second elm isolate of group IV, causes similar
symptoms in periwinkle and had Southern hybridization restriction patterns identical to the EY-MLO
(Maurer & Seemuller, 1992).
525
Group V comprises European fruit isolates. DNA
probes from the apple proliferation isolate AT crosshybridized with DNA of isolate AP and with those of the
three stone fruit isolates PARM, PPER and PSER
(Ahrens et al., 1992). However, the hybridization profiles
of the stone fruit isolates were different from that of the
apple MLOs, as observed by h a 1 restriction analysis in
this study. Genomic probes from the AT-MLO did not
hybridize with isolates of group I, 11, VI and VII (Bonnet
et al., 1990; B. Schneider, unpublished results). The
isolates of group VI are more heterogeneous than those
of groups I1 to V, which were all obtained from woody
plants and induce, depending on the group, either
virescence or non-virescence symptoms in periwinkle.
The MLOs assigned to group VI were from woody and
herbaceous hosts and cause virescence or non-virescence
diseases. The relationship of the group VI MLOs to the
virescence MLOs was also shown by Southern blot
hybridization experiments in which genomic probes from
the VAC-MLO hybridized with DNA of the ACLR- and
the PLN-V6-MLOs as well as with several isolates of
group I which mostly induce virescence symptoms
(Ahrens et al., 1992; B. Schneider, unpublished results).
Group VII, includes the only MLOs examined (SCWL
and BGWL) that are known to originate from monocots.
Although the taxonomic rank of the seven groups
established is not clear, their complexity differs considerably. The small groups I1 to V and VII include only
one organism or a few closely related MLOs. These
groupings are supported by hybridization results, host
range, and the symptoms induced. The MLOs of group
VI are more heterogeneous and remain to be further
differentiated. This is especially true of isolate SUNHP,
which may be sufficiently different from the other group
VI MLOs to form a subgroup or a group of its own. The
situation in group I is more complex because it includes
organisms which can be distinguished by their association with diseases such as aster yellows, clover phyllody,
sandal spike, or stolbur. Thus, several agents of group I
were, in the phenotypic classification of Marwitz (1990),
considered to be distinct. The slight variation in the
restriction pattern of the MLOs clustering with the
stolbur isolate STOL may indicate the presence of a
subgroup.
The M Y - , OAY- and SAY-MLOs, the three organisms of group I for which 16s rDNA sequencing data are
available, exhibit a sequence homology of at least 98.4 %.
Therefore, they appear to be closely related although
they belong to two different symptom groups. However,
in their recent contribution on the significance of 16s
rRNA data for differentiation of prokaryotes, Fox et al.
(1992) reported on bacteria which are distinguished at
the species level despite an even higher degree of sequence
homology than found in these three MLOs. The results
Downloaded from www.microbiologyresearch.org by
IP: 88.99.165.207
On: Fri, 16 Jun 2017 11:38:44
526
B. Schneider and others
of these authors indicate that a high 16s rDNA sequence
similarity is not necessarily a sufficient criterion to
guarantee species identity. To further differentiate the
MLOs, especially those of the heterogeneous groups,
additional tools such as other restriction endonucleases
for 16s rDNA analysis, specific genomic probes used in
Southern hybridization, or serological methods must be
applied. Monoclonal antibodies and polyclonal antisera
proved to be highly specific and allowed the differentiation of, for instance, the AY- and the clover phyllody
MLOs or even strains of the AY-MLO (Lin & Chen,
1985; Clark et al., 1988).
The taxonomic distance of MLOs from different
groups was also examined by comparing the 16s rDNA
sequencing data. Except for group 11, which is closely
related to group I, the homology is between 89.6 and
92.6%. This is considerably lower than between the
sequenced isolates of groups I or V. Despite the low
number of organisms on which these figures are based,
they indicate that the differences in the nucleotide
sequence are expressed in the restriction patterns. These
values also show a relatively close relationship between
the MLOs examined, which may have arisen from a
common ancestor. In other mollicutes, such as those of
the genus Mycoplasma, the differences are considerably
greater. For instance, the sequence homology of the 16s
rDNA of M . capricolum and M . hyopneumoniae is only
77.7 % (Taschke et al., 1987). These two organisms were
assigned to different phylogenetic groups by Weisburg et
al. (1989).
This work was supported by grants from the Deutsche Forschungsgemeinschaft. We thank H. Kison and R. Maurer for providing 16s
rDNA sequencing data of the ULW-, ASHY- and ACLR-MLOs. We
gratefully acknowledge L. Carraro, M. F. Clark, M. T. Cousin, F.
Dosba and R. Marwitz for supplying sources of MLOs used in this
study.
References
AHRENS,U. & SEEMULLER,
E. (1992). Detection of DNA of plant
pathogenic mycoplasma-like organisms by a polymerase chain
reaction that amplifies a sequence of the 16s rRNA gene. PhytopathOlOgy 82,828-832.
AHRENS,U., SEEMULLER,
E. & LORENZ,K. H. (1992). Genetic
differentiation of the MLOs affecting temperate fruit trees. International Organisation for Mycoplasmology Letters 2, 150.
BERTACCINI,
A., DAVIS,R. E., & LEE,I.-M. (1990). Distinction among
mycoplasma-like organisms (MLOs) in Gladiolus, Ranunculus,
Brassica, and Hydrangea through detection with nonradioactive
cloned DNA probes. Phytopathologia mediterranea 29, 107-1 13.
BONNET,
F., SAILLARD,
C., KOLLAR,
A., SEEMULLER,
E. & Bovi, J. M.
(1990). Detection and differentiation of the mycoplasma-like organism associated with apple proliferation disease using cloned
DNA probes. Molecular Plant-Microbe Interactions 3,438-443.
Bovk, J. M. & SAILLARD,
C. (1979). Cell biology of spiroplasmas. In
The Mycoplasmas, vol. 111, pp. 83-153. Edited by R. F. Whitcomb &
J. G. Tully. New York: Academic Press.
BOUVET,
A., GRIMONT,
F. & GRIMONT,
P. A. D. (1991). Intraspecific
variations in nutritionally variant streptococci : rRNA gene restriction patterns in Streptococcus defectivus and Streptococcus
adjacens. International Journal of Systematic Bacteriology 41,
483-486.
CARRARO,L., OSLER,R., REFATTI,E. & POGGIPOLLINI,C. (1988).
Transmission of the possible agent of apple proliferation to Vinca
rosea by dodder. Rivista di Patologia Vegetale S.IV, 43-52.
CLARK,M. F., DAVIES,D. L., Buss, S. L., & MORTON,A. (1988).
Serological discrimination among mycoplasma-like organisms using
polyclonal and monoclonal antibodies. Acta Horticulturae 235,
107-1 13.
DAVIS,R. E., SINCLAIR,W. A., LEE, I.-M. & DALLY,E. L. (1992).
Cloned DNA probes specific for detection of a mycoplasma-like
organism associated with ash yellows. Molecular Plant-Microbe
Interactions 5, 163-169.
J. & LEE,P. E. (1972). Transmission by dodder of sandal
DIJKSTRA,
spike disease and the accompanying mycoplasma-like organisms via
Vinca rosea. Netherlands Journal of Plant Pathology 78,2 18-228.
Fox, G. E., WISOTZKEY,
J. D. & JURTSHUK,
P. (1992). How close is
close: 16s rRNA sequence identity may not be sufficient to guarantee
species identity. International Journal of Systematic Bacteriology 42,
166-1 70.
GOBEL,U. B, GEISER,A. & STANBRIDGE,
E. J. (1987). Oligonucleotide
probes complementary to variable regions of ribosomal RNA
discriminates between Mycoplasma species. Journal of General
Microbiology 133, 1969-1974.
GRIMONT,
F. & GRIMONT,
P. A. D. (1986). Ribosomal nucleic acid gene
restriction patterns as potential taxonomic tools. Annales de I Institut
PasteurlMicrobiologie 137B, 165-1 75.
V., WILSON,V. A. & MAYALL,
B. C. (1991). Classification of
GURTLER,
medically important clostridia using restriction endonuclease site
differences of PCR-amplified 16s rDNA. Journal of General Microbiology 137,2673-2679.
HEINTZ,W. (1989). Transmission of a new mycoplasma-like organism
(MLO) from Cuscuta odorata (Ruiz et Pav.) to herbaceous plants
and attempts to its elimination in the vector. Journal of Phytopathology 125, 171-186.
HIGGINS,D. G. & SHARP, P. M. (1988). Clustal: a package for
performing multiple alignments on a microcomputer. Gene 73,
237-244.
JENSEN,D. D. (1956). Insect transmission of virus between tree and
herbaceous plants. Virology 2, 249-260.
KLEIN, M. (1970). Safflower phyllody - a mycoplasma disease of
Carthamus tinctorius in Israel. Plant Disease Reporter. 54, 735-738.
KUSKE,C. R. & KIRKPATRICK,
B. C. (1992). Phylogenetic relationship
between the western aster yellows mycoplasma-like organisms and
other prokaryotes established by 16s rRNA gene sequence. International Journal of Systematic Bacteriology 42, 226-233.
KUSKE,C. R., KIRKPATRICK,
B. C. & SEEMULLER,
E. (1991). Differentiation of virescence MLOs using western aster yellows mycoplasmalike organism chromosomal DNA probes and restriction fragment
length polymorphism analysis. Journal of General Microbiology 137,
153-159.
LAIGRET,F., GRAU,0. & Bovk, J. M. (1990). Comparison of 16s
rDNA sequences of various mollicutes. Zentralblatt fur Bakteriologie, Suppl. 20, 435-440.
LANE,D. J., PACE,B., OLSEN,G. J., STAHL,D. A., SOGIN,M. L. &
PACE,N. R. (1985). Rapid determination of 16s ribosomal RNA
sequences for phylogenetic analysis. Proceedings of the National
Academy of Sciences of the United States of America 82,6955-6959.
LEE, L.-M. & DAVIS.R. E. (1988). Detection and investigation of
genetic relatedness among aster yellows and other mycoplasma-like
organisms by using cloned DNA and RNA probes. Molecular
Plant-Microbe Interactions 1, 303-3 10.
LEEI.-M., DAVISR. E. & DEWITTN. (1990). Nonradioactive screening
method for isolation of disease-specific probes to diagnose plant
diseases caused by mycoplasma-like organisms. Applied Environmental Microbiology 56, 1471-1475.
LIM, P.-0. & SEARS,B. B. (1989). 16s rRNA sequence indicates that
plant-pathogenic mycoplasma-like organisms are evolutionarily
distinct from animal mycoplasmas. Journal of Bacteriology 171,
590 1-5906.
Lm, C.-P. & CHEN,T. A. (1985). Monoclonal antibodies against the
aster yellows agent. Science 227, 1233-1235.
Downloaded from www.microbiologyresearch.org by
IP: 88.99.165.207
On: Fri, 16 Jun 2017 11:38:44
MLO classification
MANIATIS,
T., FRITSCH,E. F. & SAMBROOK,
J. (1982). Molecular
Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold
Spring Harbor Laboratory.
MARCHOUX,~.
& GIANNOTTI,J. (1971). Interference entre deux
mycoplasmas vtgetale. Physiologie Vkgktale 9, 595-6 10.
MARCHUK,
D., DRUMM,M., SAULINO,
A. & COLLINSF. S. (1991).
Construction of T-vectors, a rapid and general system for direct
cloning of unmodified PCR products. Nucleic A c i h Research 19, 54.
MAURER,R. & SEEMULLER,
E. (1992). Genetic relatedness of European
and North American mycoplasma-like organisms from trees.
International Organisation for Mycoplasmology Letters 2, 149.
MARWITZ,R. (1990). Diversity of yellows disease agents in plant
infections. Zentralblatt fur Bakteriologie, Suppl. 20,43 1-434.
MARWITZ,R. & PETZOLD,H. (1976). Elektronenmikroskopischer
Nachweis mycoplasmaahnlicher Organismen in DelphiniumHybriden mit Blutenvergrunung und -verlaubung. Phytopathologische Zeitschrift 87, 1-1 1.
MARWITZ,R. & PETZOLD,H. (1983). Mykoplasmaahnliche Organismen als Krankheitserreger an Primeln. Gesunde Pflanze 35,
336-341.
H. & OZEL,M. (1974). Untersuchungen zur
MARWITZ,R., PETZOLD,
Ubertragbarkeit des moglichen Erregers der Triebsucht des Apfels
auf einen krautigen Wirt. Phytopathologische Zeitschrift 81, 8 S 9 1.
MARWITZ,
R., PETZOLD,
H. & ROTH,L. (1979). Eine bisher unbekannte
stolburverwandte Krankheit bei feldmaSig angebauten Pflanzen von
Solanum marginatum in Ecuador. Phytopathologische Zeitschrift 95,
305-3 17.
MARWTZ,R., KUHBANDNER,
B. & PETZOLD,
H. (1987). Ubertragung
mykoplasmaahnlicher Organismen (MLO) von hexenbesenkranken
Heidelbeeren ( Vaccinium myrtillus) auf Catharanthus roseus mit Hilfe
von Cuscuta. Nachrichtenblatt der Deutschen Pflanzenschutzdienstes
39, 129-132.
McCoy, R. E., CAUDWELL,A., CHANG, C. J., CHEN, T. A.,
CHIYKOWSKI,
I. N., COUSIN,
M. T., DALE,J. L., DE LEEUW,G. T. N.,
K. J., KIRKPATRICK,
B. C., MARWITZ,
R.,
GOLINO,
D. A., HACKETT,
H., SINHA,R. C., SUGIURA,
M., WHITCOMB,
R. F., YOUNG,
PETZOLD,
527
E. (1989). Plant diseases associated
I. L., ZHU,B. M. & SEEMULLER,
with mycoplasma-like organisms. In The Mycoplasmas, vol. V,
pp.545-640. Edited by R. F. Whitcomb & J. G. Tully. San Diego:
Academic Press.
MORENO,P., LLACER,G., & MEDINA,V. (1985). Descripcion y
comparacion de varias micoplasmosis en Vinca rosea L. Anales del
Instituto Nacional de Investigaciones Agrarias, Serie Agricola, N .
Extr. 28, 287-309.
RAZIN,S. (1989). Molecular approach to mycoplasma phylogeny. In
The Mycoplasmas, vol. V, pp. 33-69. Edited by R. F. Whitcomb &
J. G. Tully. San Diego: Academic Press.
SEARS,B. B. & KLOMPARENS,
K. L. (1989). Leaf tip cultures of the
evening primrose allow stable, aseptic culture of mycoplasma-like
organisms. Canadian Journal Plant Pathology 11, 343-348.
SINCLAIR,
W. A., BRAUN,E. J. & LARSEN,A. 0. (1976). Update on
phloem necrosis of elms. Journal of Arboriculture 2, 106-1 13.
STACKEBRANDT,
E. (1991). Unifying phylogeny and phenotypic diversity. In The Prokaryotes, vol I, pp. 1947. Edited by A. Balows,
H. G. Truper, M. Dworkin, W. Harder, & K.-H. Schleifer. New
York : Springer Verlag.
TASCHKE,C., RULAND,K. & HERRMANN,
R. (1987). Nucleotide
sequence of the 16s rRNA of Mycoplasma hyopneumoniae. Nucleic
Acids Research 15, 3918.
C., WOLTERS,
J. & HERRMANN,
R. (1990). Ribosomal RNA
TASCHKE,
genes of four Mycoplasma species. Zentralblatt fur Bakteriologie,
Suppl. 20, 329-336.
WEISBURG,
W. G., TULLY,J. G., ROSE,D. L., PETZEL,J. P., OYAIZU,
H., YANG,D., MANDELCO,
L., SECHREST,
J., LAWRENCE,
T. G., VAN
ETTEN,J., MANILOFF,J. & WOESE,C. R. (1989). A phylogenetic
analysis of the mycoplasmas : basis for their classification. Journal of
Bacteriology 171, 64 556467.
WEISBURG,
W. G., BARNS,S. M., PELLETIER,
D. A. & LANE,D. J.
(1991). 16s ribosomal DNA amplification for phylogenetic study.
Journal Bacteriology 173, 697-703.
WELVAERT,
W., SAKYN,G. & LAGASSE,
A. (1975). Recherches sur les
symptomes de la virescence chez I’Hydrangea macrophylla Thunb.
Phytopathologische Zeitschrijl 83, 152-1 58.
Downloaded from www.microbiologyresearch.org by
IP: 88.99.165.207
On: Fri, 16 Jun 2017 11:38:44