* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture Note 6
Nucleic acid analogue wikipedia , lookup
Expression vector wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Gene expression wikipedia , lookup
Interactome wikipedia , lookup
Magnesium transporter wikipedia , lookup
Drug design wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Metalloprotein wikipedia , lookup
Genetic code wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Biosynthesis wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Point mutation wikipedia , lookup
Proteolysis wikipedia , lookup
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Module 6 Bioinformatics tools
Lecture 38 Analysis of protein and nucleic acid sequences
(Part-I)
Introduction-The genetic information is stored in DNA present in the nucleus and
transfer from one generation to other generation. DNA transfers the information to the
messenger RNA (mRNA) by the process of transcription. The correct transfer of
information is ensured by the complementary base pairing between nucleotide present
on DNA and mRNA. The mRNA transfer this information in the form of protein by
the process of translation. DNA is madeup of 4 different types of nucleotides (A, T,
G, C) and triplet of nucletide (codes) is responsible for coding for amino acid present
in the protein. It is made up of different types of amino acids and composition of
protein is determined by the DNA sequence (Figure 38.1). Hence, the sequence of
nucleotide bases as well as amino acid sequence of a protein has wealth of
information used to understand structure and function of the macromolecule. In the
current lecture we will discuss the analysis of protein and DNA sequence and
conclusion drawn from the sequence information.
Figure 38.1: The flow of genetic information from DNA to protein.
Joint initiative of IITs and IISc – Funded by MHRD
Page 1 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Structure of nucleic acid- Nucleotide, the building block of nucleic acid consists of
pentose sugar, base and phosphoric acid residue. Nucleotides are connected by a
covalent linkage between pentose sugar of nucleotide and phosphoric acid of the next
nucleotide (Figure 38.2). There are 5 different types of nucleobase (cytosine, uracil,
thymine, adenine and guanine) attached to the sugar through a N-glycosidic linkage.
Uracil is found in RNA whereas thymine is present in the DNA. These nucleotide are
abbreviated with the first letter of the base to write the nucleotide sequence of the
nucleic acid, such as adenine is denoted as “A”. The bases have a specificity towards
the other base to form a pair through hydrogen bonding, “A” is making 2 hydrogen
bonding to the “T” where as “G” is making 3 hydrogen bonding to the “C”. DNA is a
double helix structure with the bases present on the both starnd and sequence
information on one strand of DNA can determine the sequence of the other strand.
Figure 38.2: The structure of nucleic acid.
Joint initiative of IITs and IISc – Funded by MHRD
Page 2 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Structure of protein-Protein is made up of 20 naturally occurring amino acids. A
typical amino acid contains a amino and a carboxyl group attached to the central αcarbon atom (Figure 38.3). The side chain attached to the α-central carbon atom
determines the chemical nature of different amino acids. Peptide bonds connect
individual amino acids in a polypeptide chain. Each amino acid is linked to the
neighboring amino acid through a acid amide bond between carboxyl group and
amino group of the next amino acid. Every polypeptide chain has a free N- and Cterminals (Figure 38.3). Primary structure of a protein is defined as the amino acid
sequence from N- to the C-terminus with a length of several hundred amino acids.
The ordered folding of polypeptide
Figure 38.3: The connection between two adjacent amino acids in a polypeptide.
chain give rise to the 3-D conformation known as secondary structure of the protein
such as helices, sheet and loops. Arrangement of the secondary structure gives rise to
the tertiary structure. α-helix and β-sheet are connected via unstructured loops to
arrange themselves in the protein structure and it allows the secondary structure to
change their direction. Tertiary structure defines the function of a protein, enzymatic
activity or a nature of structural protein. Different polypeptide chains are arranged to
give quaternary structure (Figure 38.4).
Joint initiative of IITs and IISc – Funded by MHRD
Page 3 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Figure 38.4: The different levels of organization in a protein structure.
Biological Databases-In the post genomic era, nucleotide and protein sequences from
different organisms are available. It has paved the determination of secondary and 3D structure of the proteins as well. This vast amount of information is processed and
arranged systematically in different biological databases. The information present in
these databases can be used to derive common feature of a sequence class and
classification of a unknown sequence.
Primary Database- This the collection of the data obtained from the experiment such
as sequence of DNA or Protein, 3-D structure of a protein.
Joint initiative of IITs and IISc – Funded by MHRD
Page 4 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Database of nucleic acid sequences
GenBank-This is a public sequence database and it can be accessed through a web
addess http://www.ncbi.nlm.nih.gov/genbank/. The entry into the genbank is made
through a login into the database with a pre-requisite of publication of the new
sequence in any scientific journal. Each entry in the database has a unique accession
number and it remains unchanged. A sample GenBank entry can be accessed via a
link http://www.ncbi.nlm.nih.gov/Sitemap/samplerecord.html. A typical GenBank
entry has the information about the locus name, length of the sequence, type of the
molecule (DNA/RNA), nucleotide sequence of the entry.
Entrez-Entrez system is used to search all NCBI associated databases. It is a powerful
tool to peform simple or complicated searches by combining key word with the
logical operator (AND, NOT). For example, searching a protein kinase sequence in
human can be done by the following search syntax: Homo sapiens [ORGN] AND
protein kinase.
EMBL and DDBJ- EMBL is the nucleotide sequence database present at European
bioinformatics institute where as DDBJ is the DNA sequence database present at
centre for information biology, Japan. EMBL can be accessed at http://www.embl.de/
where as DDBJ canbe accessed at http://www.ddbj.nig.ac.jp/. Everyday, GenBank,
EMBL and DDBJ synchronize their nucleotide sequence and as a result searching of a
nucleotide in any of the database is sufficient.
Database of protein sequences
SWISSPROT-it is the collection of the annoted protein sequence of the swiss
instituite
of
bioinformatics
(SIB).
SWISSPROT
can
be
accessed
at
http://web.expasy.org/groups/swissprot/. The protein sequence entry in the swissprot
is manually curated and if required it is compared with the available literature.
Swissprot is part of the UniProt database and collectively known as UniProt
Knowledgebase. A ‘niceprot’ view of the entry in swissprot database are graphically
presented for better readability and hyperlinks are given for other databases as well.
NCBI protein database-It is a compilation of the protein sequence present in other
databases. The NCBI database contains the entries from the swissprot, PIR database,
PDB database and other known databases.
Joint initiative of IITs and IISc – Funded by MHRD
Page 5 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
UniProt-EBI, SIB and Georgetown university together collected the protein
information in the form of a centralized catalogue known as universal protein resource
(UniProt). It contains the information about the 3-D structure, expression profile,
secondary structures and biochemical function of the protein. UniProt consists of 3
parts: UniProt Knowledge database (UniProtKB), UniProt Reference (UniRef) and
UniProt Archive (UniPArc). As discussed before, UniProtKB is a collection from
SwissProt and TrEMBL database. UniRef is a nonredudant sequence database and it
can allow to search similar sequences. UniRef 100, UniRef90 and UniRef50 are the
three version of the database allow searching of sequences 100%, >90% and >50%
identical ot the query sequence.
Joint initiative of IITs and IISc – Funded by MHRD
Page 6 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Lecture 39 Analysis of protein and nucleic acid sequences
(Part-II)
Secondary Database-The analysis of the primary data gives rise to the development
of secondary database. Secondary structures, hydrophobicity plot and domains are
present in the various secondary databases.
Prosite-Prosite is one of the secondary biological database which contains motifs to
classify the unknown sequence into the protein family or class of enzyme. It can be
accessed with the web address http://prosite.expasy.org/. The database contains motifs
derived from the multiple sequence alignment. The quert sequence is aligned against
the multiple sequence alignment to determine the presence or absence of the motif. A
typical expression in prosite has seven amino acid positions. For examples, [EFTNA][HFDAS]-[HYT]-{ADS}-X (2)-P. This expression can be understood as follows1st position can be E, F, T, N or A
2nd position can be H, F,D,A,S
3rd position can be HYT
4th position can be any amino acid except ADS
5th and 6th position, any amino acid can follow and the 7th position will be proline.
A query sequence can be analyzed using the algorithm ScanProsite. In addition, it
may allow to search the sequence with similar pattern in SwissProt, TrEMBL and
PDB databases.
PRINTS:
Pfam: The Pfam database contains the profiles of the protein sequences and classifies
the protein families as per the over-all profile. A profile is a pattern of the amino acid
in a protein sequence and determine probability of a given amino acid. Pfam is based
on the sequence alignment. A high quality sequence alignment gives the idea about
the probability of appearance of an amino acid at a particular position and contain
evolutionary related sequences. However, in few cases a sequence alignment may
have sequences with no evolutionary relationship to each other. A critical analysis of
result from the Pfam database is necessary to draw conclusions.
Joint initiative of IITs and IISc – Funded by MHRD
Page 7 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Interpro-SwissProt, TrEMBL, Prosite, Pfam, PRINT, ProDom, Smart and
TIGRFAMS are integrated into a comprehensive signature database known as
Interpro. The results from interpro gives the output from individual databases and
allows user to compare the output considering the algorithm used in each database.
Molecular structure database
Protein Data bank (PDB)- it is the collection of the experimentally determined
crystal stuture of the biological macromolecules. It is co-ordinated by the consortium
located in Europe, Japan and USA. As of August 2013, the database contains 93043
structures which includes protein, nucleic acids, and protein-nucleic acid or proteinsmall molecule complexes (http://www.rcsb.org/pdb/home/home.do). A PDB ID or
the key word can be use to search the database. The result from the database
summarizes all information related to the structure such as crystallization condition,
reference of the journal article where the finding are published etc.
SCOP-SCOP (structural classification of protein) utilizes the basic idea that the
proteins with similar biological functions and evolutionary related with each other
must have a similar structure. The database classifies the structure of a known protein
into the families, superfamilies and fold. A protein structure belongs to a famiy if the
sequence identity must be atleast 30% over the total length of the sequence. Proteins
with structural or functional similarity but low sequence identity are classified into the
superfamilies. Whereas proteins with similar secondary structure arrangement belongs
to the fold.
CATH-Similar to SCOP, CATH classifies the protein into 4 categories: Class (C),
Architecture (A), Topology (T), and Homologous superfamily (H). A protein is
classified as Class depending on the proportion of the secondary structure elements
rather than their arrangement. There are 4 classes, helices (α-class), sheet (β-class),
helix-sheet (α/β class) and proteins with few secondary structures. The arrangement of
secondary elements in a protein structure is used for their classification within the
architecture. The connection of secondary elements is used for their classification
within the topology category. The homologous superfamily consider the presence of
similar domains in two protein structure for their classification.
Joint initiative of IITs and IISc – Funded by MHRD
Page 8 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Sequence Comparison
Homologous- Two related sequences are termed as homologous to each other. These
can be either orthologs or paralogs. The homologous protein from two different
organsism with similar functions are termed as ortholog where as homologous
protein with different protein with different function in an organism is called as
paralog.
Identitity and similarity- The ratio of identical amino acids residues to the total
number of amino acids present in the entire length of the sequence is termed as
identity (Figure 39.1). Where as ratio of similar amino acids in a sequence relative to
the total number of amino acid present is termed as similarity. The extend of
similarity between two amino acids is calculated with a similarity matrix. An
alignment between two amino acid sequences is required to calculate identity or
similarity score. In the process, two sequence are arbitrarily placed to each other and
an alignment score is calculated. This process is repeated until best score is found. In
few cases, the length of the amino acids can be enlarged or reduced by incorporating a
residue or inserting a gap (Figure 39.1).
Figure 39.1: Sequence alignment of nucleotide and protein sequences.
Joint initiative of IITs and IISc – Funded by MHRD
Page 9 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
The use of a nucleotide scoring matrix to obtain optimal alignment of two nucleotide
sequence is given in Figure 39.2. In this case, an identity matrix is relevant as the four
nucleotide will not show any similarity to each other. As given the alignment
examples, the sliding of the sequences gives different scores (3 or 7 using identity
matrix and the alignment with the best score is choosen.
Figure 39.2: Sequence alignment of nucleotide sequences.
Opposite to the nucleotides, identity matrix is not sufficient to perform alignment of
two protein sequences. Amino acids present in two sequences may have similar or
different physiochemical properties. The probability to substitute one amino acid with
other amino acids is also considered to give the score in the matrix (Figure 39.3). For
example, aspartic acid is often observed with glutamic acid but substitution of aspartic
acid with tryptophan is rare. This is due to the gentic codes of these amino acids (
aspartate and glutamic acid has only 3rd codon different) and their properties (both
aspartate and glutamic are negatively charged amino acids). In addition, the effect of
substitution on the protein structure is also been consider to provide score in the
matrix. Asparate (negatively charged) to trptophan (aromatic) will have
severe
impact on the protein structure and hence will have lower score (In the matrix given
in Figure 39.3, such a substitution will have -4 score). The most commonly used
scoring matrix are the PAM (position assisted matrix) and BLOSUM (blocks
substitution matrix). The negative value in the matrix indicate that the occurrence is
coincidental where as positive values suggest a favorable substitution. In the example
given in Figure 39.3, the two amino acid sequences are slide over to each other to
produce two alignment. Using the blosum matrix, the amino acid alignment 1 is
giving a score 65 where as amino acid alignmet 2 is giving score of 19. In this
situation, the alignment 1 is preferred over the other and be the optimal aligment for
the given two sequences.
Joint initiative of IITs and IISc – Funded by MHRD
Page 10 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Figure 39.3: Sequence alignment of protein sequences.
Joint initiative of IITs and IISc – Funded by MHRD
Page 11 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
The Alignment of two query sequences can be global or local (Figure 39.4). In global
alignment, the complete length of the protein sequences are compared to another
where as in the case of local alignment, only a part of the sequence is compared
(Figure 39.4). The global alignment is used to classify the protein into different
classes where as local alignment is used to identify the motif or domain.
Figure 39.4: Sequence alignment of protein sequences.
Joint initiative of IITs and IISc – Funded by MHRD
Page 12 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
To compare more than two sequences, multiple sequence alignment can be performed
with ClustalW. It exploits the fact that similar sequences are usually homologous.
First the pairwise alignment are carried out with the most similar sequences. Then
based on the score of pairwise alignment, all sequences are classified into different
groups. These groups are presented as multiple sequence alignment (Figure 39.5). As
ClustalW calculates the distances between different sequences, it can be use to
generate phylogenetic tree (Figure 39.6).
Figure 39.5: Sequence alignment of protein sequences.
Joint initiative of IITs and IISc – Funded by MHRD
Page 13 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Figure 39.6: A typical phylogenetic tree
HOME ASSIGNMENT
1. Go to the plasmodium falciparum genome database (www.plasmodb.org) and
down load the protein sequence with the plasmodb ID PFD0975w.
2. Identify the homologous protein from human, mouse, e.coli and neurospora.
3. Perform a sequence alignment with the clustalW and calculate the identity and
similarity score between all sequences.
4. Using the data from the sequence alignment, draw a phylogenetic tree for
PFD0975w.
Joint initiative of IITs and IISc – Funded by MHRD
Page 14 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Lecture 40 Computer Aided Drug Design
Over-view of the computer-aided drug design-Drug design and discovery is a long
process involving identification of suitable drug target, screening and selection of the
inhibitor, toxicity analysis and pharmacological analysis of the inhibitor molecule to
suit it for therapeutic purpose. The whole process of drug design and discovery
through a traditional trial-and error approach is a lengthy, time consuming and costly
process. With the evident advancement in the computational hardware and software,
most of the drug discovery
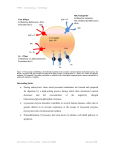
Figure 40.1: An Over-view of the different approaches used during computer-aided drug design.
steps can be performed (Figure 40.1). In a computer aided drug design approach, a
drug target is selected from the database and a 3-D structure is determined
experimentally or if the homologous structure is known then a homology model is
generated. Once the structure of the enzyme is known, active site of the enzyme is
mapped by structural comparison with known enzyme. Two approaches can be used
to design the inhibitor molecule against the enzyme, pharmacophore approach or the
docking with the random inhibitor molecules from the different chemical libraries.
Top selected inhibitor molecules can further validated in the in-silico toxicity analysis
and pharmacokinetic parameters. The best molecule can be tested further in the wet
lab experiment to validate the computational results and a series of clinical trials are
needed before allowing therapeutic applications.
Joint initiative of IITs and IISc – Funded by MHRD
Page 15 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Each step of the computer aided drug design can be performed by multiple softwares
with different algorithms. To understand the whole process of computer aided drug
design, we will take an example of an enzyme and try to design the inhibitors. This
complete process has following steps:
1. Strutural Determination of the target enzyme
A. Experimental Methods: X-ray crystallography and NMR spectroscopy are the
two methods can be used to determine the 3-dimensional structure of the target
enzyme.
I suggests to go through the following articles to get full detail of these
structure solution processes.
1. RRM-RNA recognition: NMR or crystallography…and new findings. Daubner
GM, Cléry A, Allain FH. Curr Opin Struct Biol. 2013 Feb;23(1):100-8. PMID:
23253355.
2. Protein structure determination by magic-angle spinning solid-state NMR, and
insights into the formation, structure, and stability of amyloid fibrils. Comellas G,
Rienstra CM. Annu Rev Biophys. 2013;42:515-36. PMID: 235277.
B. Homology modeling- This is a useful and fast structural solution method where
the sequence similarities between the template and the target enzyme is used to model
the 3-dimensional structure of the target enzyme. The homology modeling exploits
the idea that the amino acid sequence of a protein directs the folding of the molecule
to adopt a suitable 3-dimensional conformation with minimum free energy.
Different steps in homology modeling-Several softwares are available to perform
homology modeling of a given protein sequence (Table 40.1). Homology modeling is
a multistep process and it has following steps:
Step I : Identification of a suitable target-Identification of a suitable template
structure is the most crucial step to generate a good quality homology model. The
target sequence is blasted into the protein strucuture database (www.rcsb.org) using
PSI-Blast.
Joint initiative of IITs and IISc – Funded by MHRD
Page 16 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Step II: Sequence Alignment between target and template protein sequencetarget protein sequence is aligned against the template protein sequence using
pairwise or multiple sequence alignment (in case if more than one template proteins).
A sequence identity of more than 70% between template and target protein allows
structure prediction accurately. A sequence identity less than 30% makes structure
prediction and modeling of target protein difficult.
Step III: Model building-Template co-ordinates and the alignment information is
used to generate a 3-D structure model of the target protein. Fragment analysis and
segment analysis are two methods been used to generate the model building. The loop
modeling approach is used to model low identity amino stretch in the target protein.
Step IV: Energy minimization-The modeled structure is energy-minimized to obtain
the most stable 3-D conformation of the protein.
Step V: Structure validation-The 3-D model of the protein is validated by
Ramchandran Plot, Procheck,Verify-3D, Errat Plot. Struture validation can be
performed
by
the
structure
analysis
and
validation
(SAVS)
server
http://nihserver.mbi.ucla.edu/.
Table 40.1: Table of selected software for homology modeling.
Softwares
RaptorX
ModPipe
Biskit
SCRWL
TASSER-Lite
ProModel
LOMETS
I-TASSER
Modeller
ProSide
Prime
The utility of the software
The software is developed by Xu Group. Latest version has
four module. It is available as a software and a web service.
It is a complete automated software. It is free and a open
source software.
It is free and open source and developed by the institute
Pasteur.
The software is developed by the dunbrack lab.
It can be use to model and target protein with a sequence
identity more than 25% to the template.
Homology modeling from selected template or user provided
template. It can allow to mutation, excision, deletion etc in the
target protein.
Online web service for protein structure modeling.
Web based service for protein structure and function
prediction.
Free and one of the most popular software for homology
modeling of the target protein.
It predicts the side chain conformation.
It is a fully integrated protein structure prediction software.
Joint initiative of IITs and IISc – Funded by MHRD
Page 17 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
2. Design of the inhibitor molecules
Pharmacophore modeling-This approach is more relevant when the 3-D structure or
homology model of an enzyme is not known but the substrate or the ligand is known.
A pharmacophore is a spatial arrangement of the functional group present on the
ligand needed for the binding. To determine the pharmacophore, a series of ligand
molecules are superimposed so that similar groups come together. The common
functions are identified and categorized. The functional groups present in the ligand
molecule are hydrogen bond acceptor, donor, aromatic ring system, hydrophobic and
hydrophilic area etc (Figure 40.2). In the screening process, each molecule from the
database is fitted into the pharmacophore model and the quality of agreement is
assessed with a score. The program for pharmacophore modeling and screening are
catalyst, galahad, MOE and Phase.
Figure 40.2: Pharmacophore with the different functional groups.
Joint initiative of IITs and IISc – Funded by MHRD
Page 18 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
3. Collection of the inhibitor molecules-A list of selected database of ligand is given
in Table 40.2. For most of these database, either keyword or the chemical structure
can be used to search the database. The molecules from these database can be
downloaded in the 2-D or 3-D conformation.
Table 40.2: List of selected databases for ligand.
Database
Zinc Database
ChEMBL
Chemspider
Drug Bank
PubChem
Structural
Database
(CSD)
GPCR Ligand Library
Dictionary of Natural
Products
ChemBank
ChEBL
KEGG DRUG
The type of the ligand collection
Collection of commercially available small molecules.
Database of small molecules.
Collection of small organic molecules
A searchable collection of Drug Molecules.
Database of small molecules.
Database of 3-D structure of small molecule determined
by x-ray crystallography.
Ligands of GPCR
Database of Natural Products
Database of small molecules.
Database of small molecules.
Drug Database
4. Docking-A list of molecular modeling and docking software are given in the
Table 40.3.
Different steps in docking protocol: We will take the example of Autodock to
understand different steps of docking. Autodock 4.1 is one of the most popular
docking softwares. It has following steps to perform docking of a small moleculesStep 1 and 2: Preparation of Macromolecule and Ligand for AutoDock-Step 1
and 2 are required to give the target and inhibitor molecule suitable environment for
optimal docking. This step also allows to define the number of bonds can be made
rotable for ligand to adopt suitable conformation for fitting within the binding pocket.
Step 3: Preparation of Grid Parameter file-This step allow to select the active site
through drawing a grid of suitable size to define the space where a ligand molecule
will be docked.
Step 4: Preparing the docking parameter files- This step allow to define the energy
parameters and other docking parameters.
Step 5: Running of the docking
Joint initiative of IITs and IISc – Funded by MHRD
Page 19 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
Step 6: Analysis of Docking results-Once the docking is over, apart from the free
energy parameters, docked conformation of the ligand can be analyzed to understand
the result.
Table 40.3 : Selected List of different softwares for docking and molecular modeling
Software
AutoDock
DOCK
DOT
FADE
FlexiDock
FlexX
FTDock
Glide
Gold
GRAMM
Molegro
Docker
The utility of the software
This is a automated docking tools. Autodock is most suitaed
for docking protein and small molecule.
This software is most suited to generate protein-protein
docking and protein-DNA complexes.
It can be use to dock macromolecule to any other molecule of
any size.
FADE is used for the molecular modeling of the protein
structure.
It is used for docking of protein and small molecule.
FleXX is used to generate the protein-ligand complex.
FTDock is used to generate protein-protein or protein-DNA
complex by rigid body docking algorithm.
Glide can be use for the protein and ligand docking.
It can be used for the protein and ligand docking.
It is used to generate protein-protein or protein-DNA complex
by rigid body docking algorithm.
Virtual It can be used to predict protein-ligand interaction.
Relevance of the docking result- There are multiple approaches to understand the
relevance of docked conformation of a ligand molecule.
A. Docking against homologous host protein- A ligand molecule can be docked
against a homologous protein from the
host and the energy parameters can be
calculated. A significant difference may give confidence that the ligand molecules
will not bind to the host protein.
B. Comparison with the substrate molecule-To correlate the free energy value with
the binding constant of the ligand, a comparison with the substrate molecule can be
performed. A substrate molecule can be docked against target protein and the energy
parameters can be calculated and used for the comparison purposes to in-directly
understand the binding affinity of the ligand molecule.
Joint initiative of IITs and IISc – Funded by MHRD
Page 20 of 21
NPTEL – Biotechnology – Bioanalytical Techniques and Bioinformatics
5. In-silico toxicity prediction- The list of different softwares for toxicity prediction
can be accessed at weblink http://www.click2drug.org/directory_ADMET.html. Most
of the toxicity prediction software or web server either gives possibility of drawing
the chemical structure or use the smiles of the ligand molecule to predict the toxicity
in cell or animal based system. They also predict the carcinogenic and mutagenic
potentials of the ligand in different systems such as cells, mouse, rat etc.
HOME ASSIGNMENT
1. Go to the plasmodium falciparum genome database (www.plasmodb.org) and
down load the protein sequence with the plasmodb ID PFD0975w.
2. Identify the suitable template and perform homology modeling to prepare the
3-D model of the PFD0975w.
3. Search similar molecules to the ATP molecule from the Zinc Database
(http://zinc.docking.org/). Download the molecules.
4. Perform docking of these molecules on the 3-D model of PFD0975w with the
help of Autodock 4.1.
Joint initiative of IITs and IISc – Funded by MHRD
Page 21 of 21