* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 1.PtII.SNPs and TAS2R38.v3
Point mutation wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Genomic library wikipedia , lookup
Molecular cloning wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Epigenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Deoxyribozyme wikipedia , lookup
History of genetic engineering wikipedia , lookup
Helitron (biology) wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Microsatellite wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Microevolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
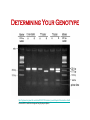
Using Single Nucleotide Polymorphism (SNP) to Predict Bitter Tasting Ability Part II: ! Digestion and Analysis of an Amplified Region of the Bitter Taste Receptor TAS2R38 Gene In The Last Lab: •! You sampled some of your own DNA and amplified a short region of chromosome 7. •! This region codes for part of TAS2R38 gene, bitter taste receptor gene. In Today’s Lab: •! You will digest some of your amplified PCR product with the restriction enzyme, Hae III •! And use agarose gel electrophoresis to determine your genotype •! You will also taste a sample of PTC paper and determine your phenotype •! You will compare the results of your genotype with those of your phenotype – record on board •! Finally, you will use class data to determine if the class is in Hardy-Weinberg equilibrium Digestion of TAS2R38 With Restriction Enzyme, Hae III Agarose Gel Electrophoresis Digesting PCR Sample with HAE III Get your PCR tube from your instructor or TA (need your code) •! Transfer 16 µl (0.016 ml) of your PCR product (amplified DNA) to a flat top PCR tube •! Label this tube “D” for “digested” and add your code. •! This DNA sample will be digested with the restriction enzyme, HaeIII •! Ask your instructor or TA to add 1µl (0.001 ml) of the restriction enzyme, HaeIII, to the “D” tube •! Place this tube in the thermal cycler set for 37oC and incubate for at least 30 minutes. Label your original PCR tube with a “U” for undigested; leave this sample on ice while the other sample is incubated with HaeIII. ! http://highered.mcgraw-hill.com/olc/dl/120078/bio37.swf http://highered.mcgraw-hill.com/sites/0072437316/student_view0/chapter16/animations.html# (choose restriction endonucleases) Agarose Gel Electrophoresis of Amplified PCR Products •!You will share an agarose gel (2%) with 2 other people •!Load both the undigested and digested samples in separate wells of the gel •!Undigested - 10 µl •!Digested - 16 µl •! Also load the Marker, a 100bp ladder, in both end lanes of the gel •! http://www.sumanasinc.com/webcontent/animations/content/gelelectrophoresis.html •! http://www.wellesley.edu/Biology/Concepts/Html/gel.html Agarose Gel Run the gel for 30-40 minutes at 130 Volts; remove gel and stain it with ethidium bromide Staining DNA - Ethidium Bromide •!Bring your gel to your TA who will transfer it to the plastic container and add ethidium bromide stain to the DNA bands •!Ethidium bromide intercalates between the base pairs of the DNA ladder •!Stain the gel ~20 minutes •!Destain ~ 10-15 minutes •!View gel using UV light to make the bands fluoresce •!Analyze photo; attach it to your worksheet Determining Your Genotype http://highered.mcgraw-hill.com/sites/0072437316/student_view0/chapter16/animations.html# Choose RFLP-restriction fragment length polymorphism Determining your PTC Phenotype •! •! •! •! Taste the Control paper first (no PTC) Taste the PTC paper Place strip in your mouth for a few seconds Remove it and wait; it takes a few seconds to taste PTC •! Record both your genotype and phenotype on board •! You will need these results to complete the post-lab assignment Class Results Genotype Phenotype Phenotype strong taster weak taster Phenotype non-taster TT Tt tt * Record the class results on your worksheet. Worksheet •! Do sample calculations •! Calculate expected allele and genotype frequencies using the sample data given: •! T/T: 15 •! T/t: 50 •! t/t: 35 •! Do Chi square calculation to determine if the sample population is in HardyWeinberg equilibrium http://www.pbs.org/wgbh/nova/sciencenow/0302/01.html (12minutes) Personal DNA Testing Next Lab DUE: C-fern Lab Report Pre-lab on RPI-LMS: Genetics & Medicine Simulation Lab: Sickle Cell Alleles Quiz: Using SNPs to Predict Bitter Tasting Ability •! How well does TAS2R38 genotype predict PTC-tasting phenotype? •! What does this tell you about classical dominant/recessive inheritance? The presence of a T allele generally predicts tasting, although heterozygotes are more likely to be weak tasters. Even in a relatively simple genetic system such as PTC tasting, one allele rarely has complete dominance over another. This experiment examined only one of several mutations in the TAS2R38 gene that influence bitter tasting ability. Variability in taste perception is likely affected by processing in the brain, which involves numerous other genes. (E.G., There are about 25-30 genes that code for bitter taste receptors. Also the genes for three of the cranial nerves involved in taste perception.) Thus, perception of bitter tasting substances like PTC is complex and does not exactly follow the simple Mendelian inheritance pattern. The forward primer incorporates part of the HaeIII recognition site, GGCC. How is this different from the sequence of the human TAS2R38 gene? Mismatch between forward primer & alleles Formation of HaeIII Recongition Site in Amplified PCR Product http://www.pbs.org/wgbh/nova/body/tongue-taste.html Making sense of SNPs and the Albuterol Story http://www.genome.gov/Pages/Education/DNADay/ TeachingTools/MakingSNPsMakeSense.html