* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Proteins
Artificial gene synthesis wikipedia , lookup
Expression vector wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Gene expression wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Interactome wikipedia , lookup
Peptide synthesis wikipedia , lookup
Western blot wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Point mutation wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Homology modeling wikipedia , lookup
Metalloprotein wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Biosynthesis wikipedia , lookup
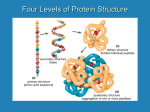
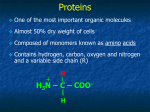
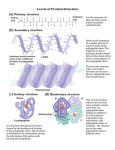
Proteins Function and Structure Proteins • more than 50% of dry mass of most cells • functions include – structural support – storage, transport – cellular communication – movement – defense against foreign substances (immunity) - enzymatic reactions Cell communication Think of conditions or diseases where malfunctioning proteins are responsible? Structure of Proteins • Monomer: amino acid • 20 different a.a. used in cells • Polymer of amino acids-->polypeptide Complex of >1 polypeptides-->protein (a protein often refers to the functional entity) Amino Acid Structure • Organic molecules with – Amino end ? – Carboxyl end ? – Central -carbon – Distinct side chain (or R group) bonded to -carbon LE 5-UN78 carbon H+ What happens to the end groups in a cellular environment? Amino group Carboxyl group How are 20 amino acids be different from each other? R groups are unique Note: R groups- aka side chains LE 5-17a Amino acids Memorize structure Glycine (Gly) Alanine (Ala) Valine (Val) Leucine (Leu) Isoleucine (Ile) Nonpolar Methionine (Met) Phenylalanine (Phe) Tryptophan (Trp) Proline (Pro) LE 5-17b Polar Serine (Ser) Threonine (Thr) Cysteine (Cys) Tyrosine (Tyr) Asparagine (Asn) Glutamine (Gln) LE 5-17c Acidic Basic Electrically charged Aspartic acid (Asp) Glutamic acid (Glu) Lysine (Lys) Arginine (Arg) Histidine (His) • Amino acids – linked together through peptide bonds • Draw dipeptide bond showing bond • Polypeptides range in length – a few a.a. to > thousand • Each polypeptide has unique linear sequence of amino acids Protein Conformation • Helices, coils, pleats • Sequence of amino acids determines 3-D conformation--> function • Depicted in ribbon and space-filling models LE 5-19 Groove A ribbon model Groove A space-filling model Four Levels of Protein Structure • Primary structure (1o) – unique sequence of amino acids, like letters in a word • Secondary structure (2o) – -helices and -pleated sheets Stabilized by H-bonds • Tertiary structure (3o) – determined by interactions among various side chains (R groups) • Quaternary structure (4o) – Multiple polypeptide chains forming a functional protein LE 5-20a 1o structure Amino end Amino acid subunits Gly Ser Tyr Phe…. Carboxyl end Four Levels of Protein Structure • Primary structure (1o) – unique sequence of amino acids, like letters in a word • Secondary structure (2o) – - -helices and -pleated sheets Stabilized by H-bonds between amino and carbonyl groups Creates 3-D conformation • Tertiary structure (3o) – determined by interactions among various side chains (R groups) • Quaternary structure (4o) – Multiple polypeptide chains forming a functional protein LE 5-20b 2o structure pleated sheet Amino acid subunits helix Four Levels of Protein Structure • Primary structure (1o) – • Secondary structure (2o) – • unique sequence of amino acids, like letters in a word -helices and -pleated sheets Stabilized by H-bonds Tertiary structure (3o) - determined by bonds between side chains (R groups) often between linearly distant amino acids -ionic bonds, disulfide bonds, van der Waals forces, H-bonds - creates to 3-D conformation • Quaternary structure (4o) – Multiple polypeptide chains forming a functional protein LE 5-20d Hydrophobic interactions and van der Waals interactions Polypeptide backbone Hydrogen bond Disulfide bridge Ionic bond Cysteines form disulfide bonds. Look at R group of cysteine to see why. Four Levels of Protein Structure • Primary structure (1o) – • Secondary structure (2o) – • unique sequence of amino acids, like letters in a word -helices and -pleated sheets Stabilized by H-bonds Tertiary structure (3o) - determined by bonds between side chains (R groups) often between linearly distant amino acids -ionic bonds, disulfide bonds, van der Waals forces, H-bonds - contributes to 3-D conformation • Quaternary structure (4o) – Multiple polypeptide chains forming a functional protein How many of you have or are singing in a choir? How many in the group? Play on a team? How many on the team? Worked in theater? With how many others? Could you have accomplished the group’s goal alone? LE 5-20e Polypeptide chain Chains Iron Heme Complex of polypeptide subunits Collagen Chains Hemoglobin LE 5-20 pleated sheet +H 3N Amino end Amino acid subunits helix Significance of Protein Conformation • Small change in 1o structure – can change protein’s conformation and function • Example – Sickle-cell disease • an inherited blood disorder-->anemia Caused by single amino acid substitution in hemoglobin LE 5-21a Normal RBC Sickled RBC 10 µm Normal cells are full of individual hemoglobin molecules, each carrying oxygen. 10 µm Fibers of abnormal hemoglobin deform cell into sickle shape. LE 5-21b One Amino Acid Substitution: Huge Effect! Sickle-cell hemoglobin Normal hemoglobin Primary structure Val His 1 2 Leu Thr 3 4 Pro Glu 5 6 Secondary and tertiary structures 7 subunit Quaternary Normal hemoglobin structure (top view) Primary structure Secondary and tertiary structures Molecules do not associate with one another; each carries oxygen. His Leu Thr Pro Val Glu 1 2 3 4 5 6 7 Exposed hydrophobic region subunit Quaternary structure Val Function Glu Sickle-cell hemoglobin Function Molecules interact with one another to crystallize into a fiber; capacity to carry oxygen is greatly reduced. Environment Affects Protein Structure & Function ? pH salt concentration temperature other environmental factors Extreme conditions cause unraveling of protein structure:denaturation LE 5-22 Caused by, for example, high temperature (100oC) Denaturation Normal protein Denatured protein Renaturation Lowered Temp (37oC) Proper Protein-Folding • Chaperonins – protein complexes that assist in the proper folding of other proteins LE 5-23a Cap Hollow cylinder Chaperonin (fully assembled) LE 5-23b Model Polypeptide Steps of Chaperonin Action: An unfolded polypeptide enters the cylinder from one end. Correctly folded protein The cap attaches, causing the cylinder to change shape in such a way that it creates a hydrophilic environment for the folding of the polypeptide. The cap comes off, and the properly folded protein is released. Techniques to Determine Protein Structure • X-ray crystallography (need to make protein crystals) • Nuclear magnetic resonance (NMR) spectroscopy (not dependent on making protein crystals) How is the sequence of proteins determined? -encoded in DNA - two step process to decode 1. DNA is transcribed into mRNA 2. mRNA is translated into polypetide More later