* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Abstract
Artificial gene synthesis wikipedia , lookup
Gene expression wikipedia , lookup
List of types of proteins wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Silencer (genetics) wikipedia , lookup
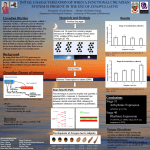
Field: Biology/Life Science Session Topic: RNA in Systems Biology /Analysis and Synthesis of Mammalian Circadian Clocks Speaker: Hiroki R. Ueda/RIKEN The logic of complex and dynamic biological networks is difficult to elucidate without (1) comprehensive identification of network structure, (2) prediction and validation based on quantitative measurement and perturbation of network behavior, and (3) design and implementation of biological networks driven by the same logic as the original network. Mammalian circadian clock system is such a system consisting of complexly integrated regulatory loops and displaying the various dynamic behaviors including 1) endogenous oscillation with about 24-hour period, 2) entrainment to the external environmental changes (temperature and light cycle), 3) temperature compensation over the wide range of temperature, and 4) synchronization of multiple cellular clocks against the inevitable molecular noise. To elucidate complex structure and dynamic behavior of mammalian circadian clock, we comprehensively identify the transcriptional regulatory circuits composed of 20 transcription factors, and three type of DNA elements including “morning” element (Bmal1/Clock Binding element, E-box/E’-box), “day” element (DBP/E4BP4 binding element, D-box) and “night” element (RevErbA/ROR binding element, RREs) (Figure 1). The following quantitative measurement and perturbation of clock circuits revealed that E-box/E'-box regulation represents a topological and functional vulnerability in mammalian circadian clocks (Figure 2), and also found the interesting property of peripheral circadian clocks. In this symposium, we will also report a current progress in the synthesis of transcriptional circuits underlying mammalian clock and discuss the logic governing this complex and dynamic biological networks. References 1. Ueda, H.R. et al. A transcription factor response element for gene expression during circadian night. Nature 418, 534-539 (2002). 2. Ueda, H.R. et al. System-level identification of transcription circuit underlying mammalian circadian clocks. Nat. Genet. 37, 187-192 (2005). 3. Sato T K, et al. Feedback repression is required for mammalian circadian clock function. Nat Genet 38:312-9 (2006). URL http://www.cdb.riken.jp/jp/02_research/0202_creative20.html Figure 1. Transcriptional circuits underlying mammalian circadian clocks. Figure 2. Circadian rhythms in wild-type (a) and mutant cells (b). Mutant cells are impaired in E-box/E’-box regulation. Each colored line indicates the single-cellular transcriptional activity while black line indicate the averaged transcriptional activity.