* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Endosymbiosis: The Evolution of Metabolism
Nucleic acid analogue wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Biosynthesis wikipedia , lookup
Transposable element wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Molecular ecology wikipedia , lookup
Genetic code wikipedia , lookup
Citric acid cycle wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene regulatory network wikipedia , lookup
Gene expression wikipedia , lookup
Mitochondrial replacement therapy wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Community fingerprinting wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Non-coding DNA wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Mitochondrion wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Molecular evolution wikipedia , lookup
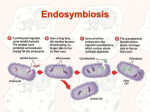
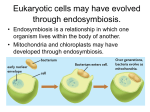
Endosymbiosis: The Evolution of Metabolism In eukaryotic cells: Why is oxidative phosphorlylation (most of ATP synthesis) done in the mitochondria? Why is the light reaction of photosynthesis done in chloroplasts? Chloroplasts and mitochondria are peculiar organelles. They have double membranes and their own circular DNA molecules. They also reproduce independently of the cell. In 1965, Dr. Lynn Margulis proposed the theory of Endosymbiosis. She claimed that mitochondria and chloroplasts are actually descendants of ancient bacteria that were captured by the ancestors of eukaryotic cells more than a billion years ago. - Endosymbiosis is clearly a crazy theory, isn’t it? - What is the evidence? Margulis presented several types of physical evidence to support her theory. 1) Mitochondria and chloroplasts have their own DNA 2) Mitochondria and chloroplasts reproduce independently of the cell 3) Mitochondria and chloroplasts have membranes that are not connected to other cellular membrane systems 4) Cases of Endosymbiosis can be found in present day organisms such as Chlorella alga inside of Paramecium This argument was not sufficiently persuasive and most scientists rejected the Endosymbiosis theory when it was first presented. However, in the early 1980’s Dr. Margulis was able to use DNA sequence analysis (bioinformatics) methods to validate the endosymbiosis theory. She compared gene sequences between mitochondria and bacteria; and also between chloroplasts and blue-green algae (photosynthetic bacteria). You can re-create her experiments using sequences from GenBank and the BLAST sequence similarity tool at the NCBI website (see the Sequence Similarity Tutorial in Appendix B). Start with genes from the mitochondria. What genes are in a mitochondrion? It has a small genome – just 16,000 bases in humans with about 20 genes. Which of these genes are likely to remain essentially unchanged over a few billion years? Respiration - specifically the Krebs cycle is highly conserved. succinate dehydrogenase is a good choice [Note: It is easier to find distant similarities if we use protein sequences rather than DNA sequences to make similarity searches.] So, use a web browser to get the succinate dehydrogenase gene sequence for human mitochondria from Genbank. GenBank is an online database which contains all known DNA and protein sequences. It is run by the NCBI (National Center for Biotechnology Information), which is part of the US National Library of Medicine at the NIH: http://www.ncbi.nih.gov Or just do a Google search for “GenBank”. Q uickTim e™ and a TI FF ( LZW) decom pr essor ar e needed t o see t his pict ur e. Who owns GenBank and the data in it? We all do – both GenBank and the sequences in it are paid for by US tax dollars allocated by congress for scientific research at the NIH and Universities across the country. Other sequences are contributed by scientists from all over the world, as a requirement for publishing articles in respected Journals. Europe and Japan maintain their own DNA sequence databases (EMBL and DDBJ), but they exchange sequences with GenBank every day, so that all three databases contain all of the same information. GenBank also includes data imported from other databases such as SwissProt and PIR. The proteins records from SwissProt are particularly valuable because they have more thorough and extensive annotation, and they have much less redundancy (duplication) than the rest of GenBank Open the NCBI website and type your search terms directly in the “Search” box near the top of the page. Use the pulldown menu to set the search for Protein. It may take a while to locate the correct succinate dehydrogenase gene if you try to search GenBank with text terms: [“succinate dehydrogenase” + human + mitochondria + subunit 2] GenBank is very big (over 2 million proteins in 2004), and the gene names are not very well organized. The Entrez search engine is very powerful, but not terribly easy to use. One shortcut is to use the locus name: DHSB_HUMAN, or the accession number: P219912. QuickTime™ and a TIFF (LZW) decompressor are needed to see this picture. Once you find the human mitochondrial succinate dehydrogenase gene, click on the link and have a look at the GenBank record. There is a LOT of information here including keywords, pointers to other GenBank records and cross-references in other databases. LOCUS DEFINITION ACCESSION VERSION DBSOURCE KEYWORDS SOURCE ORGANISM REFERENCE . . . P21912 280 aa linear PRI 25-OCT-2004 Succinate dehydrogenase [ubiquinone ] iron-sulfur protein, mitochondrial precursor ( Ip) (Iron-sulfur subunit of complex II). P21912 P21912 GI:20455488 swissprot : locus DHSB_HUMAN, accession P21912; class: standard. extra accessions:Q9NQ12,created: May 1, 1991. sequence updated: Feb 28, 2003. annotation updated: Oct 25, 2004. xrefs : gi: 665924, gi: 665925, gi: 773298, gi: 773300, gi: 773291, gi : 773292, gi: 773293, gi: 773294, gi: 773295, gi: 773296, gi: 773297, gi: 5263031, gi: 8979800, gi: 33871966, gi: 14043765, gi: 220069, gi: 220070, gi: 180916, gi: 180917, gi: 7431697 xrefs (non-sequence databases): HSSPP07014, GenewHGNC :10681, ReactomeP21912, MIM 185470, MIM 115310, MIM 171300, MIM 171350, GO0005739, GO0009060, GO0006099, InterProIPR006058, InterProIPR001450, InterProIPR004489, InterProIPR001041, InterProIPR009051, PfamPF00111, TIGRFAMsTIGR00384, PROSITEPS00197, PROSITEPS00198 2Fe-2S; 3Fe-4S; 4Fe-4S; Disease mutation; Electron transport; Iron; Iron-sulfur; Metal-binding; Mitochondrion; Oxidoreductase ; Transit peptide; Tricarboxylic acid cycle. Homo sapiens (human) Homo sapiens Eukaryota ; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi ; Mammalia ; Eutheria; Primates; Catarrhini ; Hominidae ; Homo. 1 (residues 1 to 280) Use the Display pulldown menu to show just the amino acid sequence of the protein in “FASTA” text format. Copy the amino acid sequence. Now go to the NCBI BLAST web page and choose “Protein-Protein BLAST.” Paste the DHSB_HUMAN amino acid sequence into the Search box. Choose “swissprot” from the Choose Database pulldown menu and click the nice big _BLAST!_ button. Q uickTim e™ and a TI FF ( LZW) decom pr essor ar e needed t o see t his pict ur e.