* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Poster
Nucleic acid analogue wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Gene expression wikipedia , lookup
Citric acid cycle wikipedia , lookup
Western blot wikipedia , lookup
Point mutation wikipedia , lookup
Microbial metabolism wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Peptide synthesis wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Magnetotactic bacteria wikipedia , lookup
Protein structure prediction wikipedia , lookup
Proteolysis wikipedia , lookup
Metalloprotein wikipedia , lookup
Genetic code wikipedia , lookup
Biochemistry wikipedia , lookup
Epitranscriptome wikipedia , lookup
Amino acid synthesis wikipedia , lookup
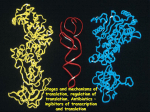
GatCAB: A Potential Target for Bacterial Destruction Greenfield High School SMART Team: Morgan Borchardt, Srinidhi Emkay, Hannah Flees, Amanda Miller, Joey Krasovich, Robin Sandner, Phat Nguyen, Panfua Thao, Tammy Tian, Tania Alvarez, Haleigh De Smet, Vivian Ramirez Teacher: Julie Fangmann Mentor: Martin St. Maurice, Ph.D., Marquette University Abstract Staph infection is caused by the bacteria Staphylococcus aureus, which have become increasingly resistant to a broad spectrum of antibiotics. New ways to combat these bacteria are needed. The Greenfield High School SMART (Students Modeling A Research Topic) Team is modeling the enzyme GatCAB using 3D printing technology. GatCAB is found in certain bacteria and archaea and could be a target for new antibiotics. During protein synthesis, ribosomes bring together aminoacylated-tRNA molecules to form proteins needed for survival. Some bacteria have tRNAs that always have an incorrect amino acid attached. Staphylococcus aureus contains such misacylated tRNA molecules with aspartate where asparagine should be attached. GatCAB’s three proteins (GatA, GatB, and GatC) correct these misacylations. GatA’s active site produces ammonia, which travels through a tunnel leading to GatB. GatC holds GatA and GatB together. GatB’s hinge recognizes and binds to the T loop of the misacylated tRNA, so GatCAB will not correct a tRNA that should have aspartate. The aspartate on the tRNA enters GatB’s active site, where the aspartate reacts with ATP and the ammonia from GatA to form asparagine, ensuring the correct amino acid is on the proper tRNA. If the misacylated tRNAs were left uncorrected, protein synthesis would be severely disrupted, and the bacteria would die. If scientists produce a drug to prevent GatCAB from fixing the misacylated amino acids, the world would have a new weapon to fight antibiotic resistant bacteria. MRSA and Antibiotic Resistance •Staphylococcus aureus causes staph infection •While antibiotics (such as penicllin and amoxicillin,)can be used to kill bacteria, some strains have become antibiotic resistant •MRSA (Methicillin-Resistant Staphylococcus aureus) is a strain resistant to a variety of antibiotics •Incidences of MRSA have risen over the years (see graph) •New ways to kill the bacteria are needed… How GatCAB Fixes Asp-tRNA Misacylations III. Ammonia Travels Through the Tunnel GatCAB with Asp-tRNA GatA GatA’s active site The Tunnel GatC ATP in GatB’s active site GatB Misacylated aspartate tRNA-Asp tRNA’s T loop GatB’s hinge tRNA’s anticodon One of the misacylations that occurs in some bacteria, such as Staphylococus aureus, occurs when an aspartate (Asp) is placed where an asparagine (Asn) is supposed to be. The misacylated Asp-tRNA can be fixed by GatCAB, as explained here. Pdb file: GatCAB_tRNA.pdb I. GatB hinge recognizes and binds to tRNA-Asp’s T-loop II. Formation of Ammonia in GatA’s Active Site pdb file: GatCAB_tRNA.pdb Protein Synthesis: The Basics Protein synthesis is the process in which proteins are made. Proteins are composed of chains of amino acids bonded together. Molecules called tRNA (transfer RNA) carry single amino acids to a ribosome, where these amino acids get attached to one another in a sequence based on the genetic information in the mRNA (messenger RNA). pdb file: GatCAB_tRNA.pdb The T-loop (dark purple) on AsptRNA binds to the hinge (blue) on GatB. The hinge will only bind to the specific sequence of the T loop on the misacylated tRNA. Once they bind together, the aspartate (Asp) inserts into the active site of GatB so it can be fixed. Ammonia is needed in order to change the misacylated aspartate into the appropriate asparagine (Asn). The active site in Gat A (light purple) is where the reaction takes place. The reaction (below) uses water and serine165 (gray, blue and red above) in GatA to remove the NH3+ from the asparagine. Once formed, the ammonia will need to get to Gat B’s active site. pdb file: GatCAB_tRNA.pdb GatCAB has a tunnel (orange) between GatA and GatB. The ammonia formed in GatA travels through the tunnel to GatB’s active site, where it is needed. IV. GatB’s Active Site Fixes the Misacylated Asp-tRNA pdb file: GatCAB_tRNA.pdb The misacylated aspartate (magenta) on the tRNA is already inserted into GatB’s active site, where ATP (gray, blue, and red), ammonia (not present in model), and the aspartate react to form asparagine. [This reaction is essentially the reverse of that in GatA’s active site (seen in box II.).] Finally, the appropriate amino acid is on the Asn-tRNA. V. Gat C Holds GatCAB Together pdb file: GatCAB_tRNA.pdb Image supplied by Dr. Martin St. Maurice Gat C (coral) works as a “belt” that keeps Gat A and Gat B together. Without Gat C the whole protein will fall apart and not function. The Parts of tRNA That Interact with GatCAB •The anticodon matches with a codon region on mRNA to indicate which amino acid needs to be added. •Each anticodon determines which specific amino acid is supposed to be carried by the tRNA. •A misacylated tRNA has an incorrect amino acid. •Misacylations can cause proteins to malfunction, possibly becoming detrimental to the organism. •Each tRNA also has a unique T loop, which in this case can assist in binding to GatCAB. A SMART Team project supported by the National Institutes of Health Science Education Partnership Award (NIH-SEPA 1R25RR022749) and an NIH CTSA Award (UL1RR031973). GatCAB Could Be Used As a New Method for Killing Certain Types of Antibiotic Resistant Bacteria As bacteria, such as Staphylococus aureus, become more resistant to a wide variety of antibiotics, new ways to kill these disease-causing bacteria are needed. Blocking GatCAB’s ability to fix misacylations might result in malfunctioning proteins and bacterial death. Image A shows the structures of Asp-tRNA (when n= 1) and Glu-tRNA (when n=2). In a study conducted by Jonathon L. Huot, et. al., a competitive inhibitor of GatCAB (aspartycin) was tested. Adding higher concentrations of aspartycin caused less activity by GatCAB. Other similar inhibitors have also been tested, not all with the same outcomes. Image B shows the structure of the competitive inhibitors, aspartycin (when n=1) and glutamycin (when n=2). This figure shows that as the concentrations of aspartycin [ ](or glutamycin) [ ] increase, the activity of GatCAB decreases. This shows aspartycin and glutamycin are competitive inhibitors of GatCAB. Images and data from: Huot, Jonathon L., et. al.; Mechanism of a GatCAB Amidotransferase: Aspartyl-tRNA Sythetase Increases Its Affinity for Asp-tRNAAsn and Novel Aminoacyl-tRNA Analogues Are Competitive Inhibitors; Biochemistry; 2007; 46; 13190-13198