* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download A1990CE78700001
12-Hydroxyeicosatetraenoic acid wikipedia , lookup
Duffy antigen system wikipedia , lookup
Cancer immunotherapy wikipedia , lookup
Immunocontraception wikipedia , lookup
Anti-nuclear antibody wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Immunosuppressive drug wikipedia , lookup
Molecular mimicry wikipedia , lookup
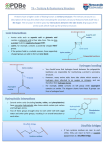
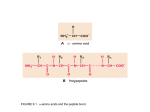
_ThIs Week’s Citation CIassIc~ ~ Wu I T & Kabat E A. An analysis of the sntjuences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complenientarity. J. Exp. Med. 132:211-50, 1970; Kabat E A & Wu T T. Attempts to locate complernentarity-deterrnining residues in the variable positions of light and heavy chains. Ann, NY Acad. Sd. 190:382-93, 1971. lDepts. Microbiol., Neurol., and Human Genet. and Develop. Coil. Physicians and Surgeons, Columbia Univ.. and Neurol. Inst.. Presbytenan Hosp., New York; Depts. Phys. and Engineer. Sin.. Northwestern Univ., Evanston. IL; and Biomath. Div., Grad. Sch. Med. Sd.. Cornell Univ.. and sloan-Kettering Inst.. New York, NY) a quantitative measure, variability: Variability = Number of These papers made use of available sequence data in 1970 and 1971 of variable regionsof light and heavy chains of ire- different amino acids at a given position/Frequency of the most common amino acid at that position. munoglobulins to predictthat the annbody-combining site was formed by hypervariable regions and that they5contained the When variability was plotted against amino acid position forthe variable region of 71) human x and I, and 7 mouse u complementarity determining residues. (The 5C1 indicates that complete and partial light chains, the sequences of which these papers have been cited in over 630 publications,( were determined by many investigators, three distinct peaks _____________________________________________ bracketed by invariant or nearly invariant residueswere seen at positions 24 to 34, 50 to56, and 89 to 97, designated as ______ hypervariable. There were insertions numbered 27A to 27F Complementarity-Determining and others around position 95 now denoted by 95A to 9SF Regions oj Antibodies based on moredala. We predicted that these, together with similar reejons in heavy chains, would form the antibody-com~ Tai Ic Wu bining site. Nowwith many more sequences Irons various species of definedspecificities and from inbred lines, the peaks Departments of Biochemistry, Molecular Biology, and Cell Biology; Biomedical Engineering in the variability plot have changed little. and Engineering Sciences and Applied Mathematics Th~i~iilowing year, we made a similar plotfor the variable Northwestern University region of 37 complete and partial heavy chains, and again three distinct peaks were present as we had expected. Within Evanston, IL 60201 a few years, our prediction was substantiated by three-dimenand sional structures ofseveral hapten-binding Fal, fragments, light Cancer Center chain dimers, and one Fv fragment, and the sin hypervari. Northwestern University Medical School Chicago, IL Cr0611 able regions were named complementarity-determining regions (CDRs). More recently, a complex of lysozyme and an September 26, 1989 antilysozyme Fab fragment was crystallized, and its three-dir 4 mensional structure was determined to 2.8 A resolution. Fifteen amino acid residues from all six CDRsand Iwo frameWhen I was at Cornell University Medical College, I read 2 two very interesting articles” by th-inA. Kabat of Columbia work residues immediately adjacent tothe CDRs madeconUniversity Collegeof Physicians and Surgeons analyzing the tact with the lysozyme molecule. Two other lysozyme-antilysozyme complexes recognized differentepitopes and again aminoacid distributions of tightchains of immunoglobulins. Since I knew very little about immunology, I wrote to Elvin all six CURs were involved.~’Currently, all X-ray structures of immunoglobulins have confirmed our prediction. asking whether I could spend some time in hislaboratory. After learning that I was also trained in engineering and The fundamental contribution of our papers is the importantprediction that antibody-combining sites are formed by applied mathematics andwas interested in mathematical bio. physics, Elvin suggested instead that we should meet once the CDRs. It therefore becomes possible to design antibodies with desired binding properties using recombinant DNA techa week to analyze the known sequence data on antibodies in more detail, initially by writing them on long strips of paper niques. Several investigators have replaced CDRs of human antibodies with those of mouse monoclonal antibodies with and later by using computers. We reasoned that the variable region of light chains ofimmunoglobulins could have random anticancer activities, In addition, variability plots have 5 also been used to tryto locate COBs in 1-cell receptors, to tieamino acid substitutions just like other proteins. However, fine residues that make contact with processed antigen and at the antibody—combining sitet many more substitutions would be needed to accommodate the vast number of dif. with 1-cell receptor 9 in the major hittocompatibility complex ferent antigens. Such amino acid variaf ions were previously class I molecules and in the class I X-ray structure,’°and to noted by Elvin as well as by Hilschmann and Craig, Putnam, identify hypes-variable regionsofvarious strains ofhuman imEdeinsass.- Fran8k.- Milstein and Pink,- and others. We defined munodeficiency viral envelope glycoproteins.” - I. Kabul K A. The paucity of species speciFic amino arid residues us the variable regions of human and mouse Bence Jones proteins and its evolutionary and genetic implications. Pt-ne. Nat. Aced. Sin. USA 57:1345-9, 1967. (Cited 25 times.) 2. . A comparison of invariant residues in the variable and constant regions of human a, bunsen )~ and mouse ii Bence Jones proteins. Pt-ssc. Nat. Aced. Sin. USA 58:229-33, 1967. (Cited 15 times.) 3. Kabat E A, Wo T T, Reid-Miller M, Perry H M & Gottesoaan K S. Sequences of proteins ofimmunological interest. Washington. DC: US Deparunent of Health and Human Services. Public Health Service, National tnststutes of Health, 1987. 804 p. (Cited 135 times.) 4. Mutt A Q, Mariuzze R A, Phillips S K V & PoIjak It J. Three dimensional structure of an antigen-antibody complex at 2.8 A resolution. Science 233:747-53, 1986. (Cited 180 times.) 5. Sheriff S, Sllvertnn K W, Padlan K A, Cohen C H, Smith-Cu S J, Fiozel B C & Davies I) R. Three-dimensional structure of an antibody-antigen complex. Plr,c. Nat. Aced. Sd. USA 84:8075-9. 1987. (Cited 35 times.) 6. PadlanEA,SllvertonEW,SheriffS,CobenGH,Smitb-GUISJ&Das’iesDR. Strucmreofanantibody-antigen complex: crystal structure of the HyHEL-lO Fab-lysozyme complex. Pine. Nat. Aca’S Sd. USA 86:5938-42, 989. 7. Jones P T, Dear P H, Foote J, Neuberger M S & Winter G. Replacing the complementarity-drtermining regions in a human antibody with those from a mouse. Nalure 321:522-5, 1986. (Cited 45 times.) 8. Becker 0 M, Patten P. Chien ‘1, Yokota T, Rshhar Z, Giedlin M, Gascoigue N R J, Goodnow C, Wolf R, Arni K & Davis M M. Variability and repertoire size of 1-cell receptor Via gene segments. Nature 317:430-4, 1985. (Cited 60 times.) 9. Pus-ham P, Lonwn C E, Lawlor t) A, Ways.! P, Holmes N, Coppin H L, Salter It U, Wan A M & Room P U. Nature of poly~~rnliisiis its HLA-A. -B, and -C molecules. Pt-nc Nat. Acad~Sin. USA 85:4(8)3-9, 198K W~J~rlthaiiFJ,S~perM A, Sam aoui u~Wti~romingerJ L & Wiley D C Structure of the human class I histocompatibility antigen HLA-A2. Nature 329:506-12, 1987. (Cited 225 times.) II. . The foreign antigen binding site and T cell recognition regions of class I itislocompatibility antigens. Nature 329:512-8, 1987. (Cited 195 times.) 18 c ©19gobylSl~l CURRENT CONTENTS~’