* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Computational Immunology An Introduction
Complement system wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Lymphopoiesis wikipedia , lookup
DNA vaccination wikipedia , lookup
Monoclonal antibody wikipedia , lookup
Psychoneuroimmunology wikipedia , lookup
Immune system wikipedia , lookup
Major histocompatibility complex wikipedia , lookup
Adaptive immune system wikipedia , lookup
Immunosuppressive drug wikipedia , lookup
Cancer immunotherapy wikipedia , lookup
Adoptive cell transfer wikipedia , lookup
Innate immune system wikipedia , lookup
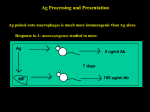
Computational Immunology An Introduction Rose Hoberman BioLM Seminar April 2003 Overview • Brief intro to adaptive immune system – B and T cells • Achieving specificity – Antibodies, TCR, MHC molecules • Maintaining tolerance to self – Clonal selection/deletion in the thymus • Paper: – Compositional bias and mimicry toward the nonself proteome in immunodominant T cell epitopes of self and nonself antigens. Innate and Adaptive • Both identify and attack foreign tissues and organisms • Have different strengths • In a constant dialogue with each other • Complement each other Innate Immunity • Recognize classes of pathogens, not a specific organism • Always respond to a pathogen in the same manner • all plants, animals, insects... have an innate immune system • example: complement binds to mannose on bacterial cell walls, flagging for phagocytosis Adaptive Immunity • Memory – enables vaccination and resistance to reinfection by the same organism • Specificity – distinguish foreign cells from self – distinguish foreign cells from one another ... the focus of this talk The Major Players • B cells – produce antibodies which bind to pathogens and disable them or flag them for destruction by the innate system • T cells – kill infected cells – coordinate entire adaptive response B cell Specificity • ImmunoGlobulin (Ig) molecules – Thousands on surface of each B cell – Ig are essentially just bound antibodies – 10^15 Ig types • Through a complicated process of DNA rearrangement ... • Each B cell’s Ig molecules recognize a unique three dimensional epitope Specificity of T cells • Each T cell has a unique surface molecule called a T cell receptor (TCR) • Through similar process of DNA splicing... • Like Ig’s, each cell’s TCRs recognizes a unique pattern (10^7 TCR types) • But a T cell epitope is a short amino acid chain (a peptide), not part of a folded protein Predicting Epitopes • Many proteins are not immunogens • Even an immunogenic protein might have only one or a few epitopes • We have millions of T and B cells, each of which recognizes only a few proteins • How can we predict epitopes? – i.e. for vaccine development, cancer treatment... Two Possible Constraints • Machinery for turning proteins into peptides – Many peptides will never even be presented to T cells • Self-tolerance – T and B cells should not attack self proteins Peptide Generation • Cytosolic proteins are degraded by a large protease complex called the proteasome • Peptides of around 8-11 a.a. are transported by TAP proteins into the ER • In the ER, a small number of peptides are bound to MHC class I molecules • These MHC-peptide complexes are shipped to the cell surface to be surveyed by T cells Peptide Generation MHC Diversity • Three loci code for MHC Locus Alleles Class I molecules and ~220 A six loci for the MHC ~110 C Class II molecules ~460 B • Most polymorphic genes DR 1,~360 in vertebrates DQ 22, 48 • Diversity is concentrated DP 20, 96 in peptide binding groove MHC-Peptide Binding TCR-MHC-Peptide Binding Learn MHC Binding Patterns • Binding databases – over 10,000 synthetic and pathogen-derived peptides – ~400 MHC I and II alleles – some qualitative affinity data – some TAP binding and T cell epitopes • Prediction methods – – – – motifs position specific probability matrices neural networks peptide threading Self Tolerance • T cells originate in the bone marrow then migrate to the Thymus where they mature • Selection of T cells through binding to common MHC-self peptides in thymus – strong binders are killed (clonal deletion) – weak binders die from lack of stimulation (clonal selection) • Remaining T cells are no longer selfreactive (with about 10 caveats) – many self-reactive T cells – danger theory Finding Immunogenic Regions of Proteins • Motivation – vaccine development – drug development for auto-immune diseases – developing techniques to co-opt the immune system for cancer therapy • Method 1: – learn to predict which peptides will be generated, transported, and bound with MHC molecules • Method 2: – learn to discriminate self from non-self and use these models to classify each possible peptide Molecular Mimicry • Protein fragment from a pathogen (or food) sometimes resembles part of a self protein • Stimulates the immune system of susceptible individuals (depending on MHC type) to attack the self protein • Can result in auto-immune disease – Shouldn’t these T cells have been filtered out? – Why isn’t the result immune ignorance? Brief Paper Overview Compositional bias and mimicry toward the nonself proteome in immunodominant T cell epitopes of self and nonself antigens Ristori G, Salvetti M, Pesole G, Attimonelli M, Buttinelli C, Martin R, Riccio P. Unigram Models Ristori... 1. Human proteome 2. Microbial proteomes (Bacteria/Viruses) We tried... 1. Human proteome 2. Pathogenic bacteria 3. Non-pathogenic bacteria unigrams.pdf Self-Reactive Protein • Multiple Sclerosis (MS) is caused by the destruction of the Myelin sheets which surround nerve cells • T cells erroneously attack the Myelin Basic Protein (MBP) on the surface of the Myelin cells • Well-studied protein; known which regions are immunogenic A Simple Self/Non-Self Predictor • For each window of size ~7-15 • Calculate the probability that the subsequence was generated by each unigram distribution • The ratio of the two gives a prediction of the degree of expected immune response • probability ratios for MBP Where to Go From Here? • Go beyond the unigram – higher level n-gram – amino acid classes – other ideas • Combine methods 1 and 2 – use to evaluate immune response dependent on an individual’s MHC alleles • Evaluation metric – classification or estimation task? • More data