* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Darren Flower - UK-QSAR
DNA vaccination wikipedia , lookup
Immunocontraception wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Monoclonal antibody wikipedia , lookup
Vaccination wikipedia , lookup
Gluten immunochemistry wikipedia , lookup
Immunosuppressive drug wikipedia , lookup
Antimicrobial peptides wikipedia , lookup
Major histocompatibility complex wikipedia , lookup
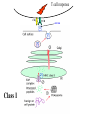
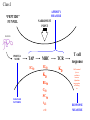
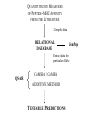
QSAR and the Prediction of T cell Epitopes Darren R Flower http://www.jenner.ac.uk/res-bio [email protected] VACCINE MARKET VACCINES 1% $5 Billion Growth in sales Vaccines: 12% yr-1 Drugs: 5% yr-1 DRUGS 15 $ 15 B $5B $ 1.75 B 5 1.75 1990 2000 2010 99% $350 Billion HUMAN VACCINES ARE MOVING FROM A MARGINAL TO A MAJOR R & D DRIVEN SECTOR AIDS ANTIBIOTIC RESISTANCE BIOTERRORISM A limited number A few innovative of vaccines targeting vaccines with major diseases blockbuster potential 1980 # of Biotech companies R &D pipeline: 100s of new vaccines 2000 10 150 IMMUNOVACCINOLOGY Vaccines induce protective immunity. Protective immunity is an enhanced adaptive immune response to re-infection. WHOLE ORGANISM attenuated SUBUNIT VACCINE EPITOPE VACCINE Delivered as recombinant protein / vector or as naked DNA + adjuvants &/or “danger signals” Types of Peptide Epitope Conformational Antibody or “B cell” Epitope Linear B cell Epitope NonConformational T cell Epitope Class I MHCs Class II MHCs all cells Professional Antigen Presenting cells Foreign and self proteins Foreign proteins 8-10 amino acids 8-20 amino acids T cell response CD8 Class I TCR PEPTIDE Class I AFFINITY MEASURE NARROWEST POINT “PEPTIDE” FUNNEL PROTEIN PROTEA SOME TAP MHC TCR IC50 IC50 KD KD CLEAVAGE PATTERNS BL50 C50 SC50 t1/2 etc. T cell response Half maximal lysis or qualitative measure (thymidine incoporation, cell killing, etc RESPONSE MEASURE PREDICTING EPITOPES Traditional Motifs: X{Y/F}XXPXXWS Frequency Matrices, Profiles “AI” Solutions: Neural Networks, HMMs, etc Epitope prediction is a chemical problem. We have taken a quantitative approach. Molecular Dynamics Quantitative Structure Activity Relationships 3D-QSAR: ComFA / CoMSIA 2D-QSAR: Free Wilson Analysis QUANTITITATIVE MEASURES OF PEPTIDE-MHC AFFINITY FROM THE LITERATURE Compile data RELATIONAL DATABASE JenPep Extract data for particular Allele QSAR CoMSIA / CoMFA ADDITIVE METHOD TESTABLE PREDICTIONS EDWARD JENNER INSTITUTE FOR VACCINE RESEARCH JENPEP Helen McSparron Martin Blythe Christianna Zygouri JENPEP VERSION 1.0 2061 T-Cell Epitopes 5848 MHC Binding data (IC50, BL50, t1/2, etc) 432 TAP Binding data ACCESS Relational Database GUI using HTML and ASP on our website: www.jenner.ac.uk/JenPep MJ Blythe, IA Doytchinova, and DR Flower. JenPep: a database of quantitative functional peptide data for immunology. Bionformatics 2002 18 434-439 JENPEP “VERSION 2.0” 3018 T cell epitopes 12210 MHC Binding data (IC50, BL50, t1/2, Kd, etc) 441 TAP Binding data 1656 B cell epitopes 300 pMHC-TCR Binding data bespoke postgreSQL relational database GUI using perl and HTML on our website: www.jenner.ac.uk/JenPep H McSparron, C Zygouri, D Taylor, MJ Blythe, IA Doytchinova, and DR Flower. JenPep+: Novel developments in quantitative immunological databases Nucleic Acids Research, commissioned Develop database system further: extend existing databases (T cell, MHC, TAP, B cell, pMHC-TCR) with new data and further retrospective analysis add new database sections: non-natural peptides and non-natural MHC mutants antibody binding whole protein antigens Host - Superantigen / Virulence Factor Binding Data Co-receptor Binding Data etc. Binding Affinity of peptides vs. Host Immunogenicity MHCs: hundreds of alleles. Each with a different peptide binding selectivity. T cell epitopes bind well to MHCs. 95% of all known T cell epitopes bind to MHC with an IC50 < 500nM. Exact T cell response is dependent on the T cell repertoire. Therefore, prediction of MHC binding is “best” option for predicting T cell epitopes. EDWARD JENNER INSTITUTE FOR VACCINE RESEARCH PREDICTING T cell EPITOPES Irini Doytchinova Christianna Zygouri PingPing Guan T - Cell Epitope Search CAVEAT our peptide sets are larger than is typical in the pharmaceutical literature the peptides themselves are physically large physical properties of peptides are extreme: multiple charges, zwitterions, huge range in hydrophobicity, etc. Sequence & thus properties are heavily biased in our peptide sets Affinity data is “poor”: multiple measurements of same peptide with orders of magnitude differences, some values are clearly wrong, mix of different standard peptides in radioligand competition assays, etc. performing a “meta-analysis”: probably many different binding modes forced into one QSAR model Predicting T cell Epitopes Using QSAR CoMFA / CoMSIA Towards the quantitative prediction of T-cell epitopes: CoMFA and CoMSIA studies of peptides with affinity to class I MHC molecule HLA-A*0201. Doytchinova, I.A and Flower, D. R. J. Med. Chem. 2001, 44, 3572-3581. Physicochemical Explanation of Peptide Binding to HLA-A*0201 Major Histocompatibility Complex. A Three – Dimensional Quantitative Structure – Activity Relationship Study. Doytchinova, I.A and Flower, D. R. Proteins, in press. FREE WILSON ANALYSIS An Additive Method for the Prediction of Protein-Peptide Binding Affinity. Application to the MHC Class I Molecule HLA-A*0201 Irini A. Doytchinova*, Martin J. Blythe and Darren R. Flower J. Proteome Research 2002, 1, 263-272. HLA-A*0201 most common allele in Caucasian population: 40% ~5x more binding data than for any other allele 152 peptides with affinity to the HLA-A2.1 Comparison of CoMFA & CoMSIA for HLA-A*0201 Training set 102 peptides CoMSIA model 9 9 8 8 predicted pIC50 predicted pIC50 CoMFA model 7 7 6 6 5 5 5 6 7 experimental pIC50 2 r = 0.694 r pred Test set 50 peptides 8 < 0.5 NC = 6 q2 = 0.480 r2 = 0.911 9 5 6 7 8 9 experimental pIC50 r = 0.783 r2pred = 0.679 NC = 5 q2 = 0.542 r2 = 0.870 Full CoMSIA Analysis of HLA-A*0201 Electrostatic Map Steric Map Hydrogen Bond Map Hydrophobic Map NC = 7 q2 = 0.683 r2 = 0.891 n = 236 ADDITIVE METHOD FOR BINDING AFFINITY PREDICTION P2 H H P4 O N H O N N H H H H P5 9 8 7 i 1 i 1 i 1 H O N N O P3 P8 O N N O P1 P6 H N N O H O O P7 pIC 50 const Pi Pi Pi 1 Pi Pi 2 HLA-A*0201: OH NC = 5 q2 = 0.337 r2 = 0.898 n = 340 P9 Amino acids contributions 1-2 Interactions 1-3 interactions How does the additive method work? YLSPGPVTV with pIC50 exp = 7.642 pIC50 = const + 1Y + 2L + 3S + 4P + 5G + 6P +7V + 8T + 9V + 1Y2L + 2L3S + 3S4P + 4P5G + 5G6P + 6P7V + 7V8T + 8T9V + 1Y3S + 2L4P + 3S5G + 4P6P + 5G7V + 6P8T + 7V9V pIC50 = 6.213 +0.304 + 0.219 – 0.164 + 0.135 + 0.013 - 0.008 + 0.096 + 0.035 + 0.263 + 0.240 – 0.015 + 0 + 0 + 0.101 + 0.075 + 0.059 + 0.102 + 0.031 + 0.044 – 0.107 + 0.046 + 0.011 + 0.008 - 0.001 = 7.700 CoMSIA & ADDITIVE METHOD ARE COMPLEMENTARY CoMSIA is “slow” but is “better” at extrapolating. ADDITIVE is very fast (analyze whole microbial genome in a few minutes) but is worse at extrapolating to peptide sequences very different to training data (missing values) Our models are not perfect but our results are at least as good as anyone else working in predicting MHC binding Trying to develop a range of “universal” models each covering a different allele A2-Supertype models CoMSIA models. parameter A*0201 A*0202 A*0203 A*0206 A*6802 n 236 63 60 54 45 q2 0.683 0.534 0.621 0.523 0.385 NC 7 8 6 12 4 SEP 0.443 0.509 0.595 0.505 0.652 SEP/affinity range % 9.9 13.6 13.0 13.3 14.8 r2 0.891 0.935 0.966 0.991 0.944 parameter A*0201 A*0202 A*0203 A*0206 A*6802 n 335 69 62 57 46 q2 0.377 0.317 0.327 0.475 0.500 NC 6 9 6 6 7 SEP 0.694 0.606 0.841 0.576 0.647 SEP/affinity range % 15.5 16.2 18.3 15.2 14.7 r2 0.731 0.943 0.963 0.989 0.983 Additive models. MHCPred: an on-line server for peptide MHC binding prediction Models: A*0101, A*0201, A*0202, A*0203, A*0206, A*0301, A*1101, A*3301, A*6801, A*6802, B*3501 www.jenner.ac.uk/MHCPred P Guan, IA Doytchinova, C Zygouri, and DR Flower. MHCPred: bringing a quantitative dimension to online prediction of MHC Binding. To be submitted FUTURE DEVELOPMENTS OF THIS WORK Make “true” predictions - design new peptides and test them experimentally Develop models for uncharacterized MHC alleles using peptides generated with Experimental Design In Progress Develop Additive Method to be descriptor based Develop “better” QSAR models using “clean” thermodynamic data from ITC and designed peptides Planned