* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Slide 1
Bimolecular fluorescence complementation wikipedia , lookup
Protein folding wikipedia , lookup
Cooperative binding wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Western blot wikipedia , lookup
Protein purification wikipedia , lookup
List of types of proteins wikipedia , lookup
Structural alignment wikipedia , lookup
Circular dichroism wikipedia , lookup
Trimeric autotransporter adhesin wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Homology modeling wikipedia , lookup
Protein domain wikipedia , lookup
Protein–protein interaction wikipedia , lookup
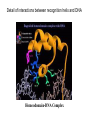
Helix-loop-Helix Motif: Simple is Not Boring! Chet Fornari, Jennifer Inlow, Larry Merkle, Barry Muhoberac, Yosi Shibberu From http://www.biologie.uni-hamburg.de/lehre/mikro/mikro402/mi4_711i.jpg The beauty, simplicity, and elegance of the helix-loop-helix protein structural motif makes it an excellent instructional tool. (A) The carboxyl-terminal helix (red) is called the recognition helix because it participates in sequence-specific recognition of DNA. (B) This helix fits into the major groove of DNA, where it contacts the edges of the base pairs. From Molecular Biology of the Cell. 3rd ed. Alberts, Bruce; Bray, Dennis; Lewis, Julian; Raff, Martin; Roberts, Keith; Watson, James D. New York and London: Garland Publishing; c1994. Detail of interactions between recognition helix and DNA Homeodomain-DNA Complex A common, fundamental structural motif of diverse proteins that interaction with DNA, such as transcription factors, restriction enzymes, and topoisomerases: Examples: bacteria yeast drosophila mammals Two aspects to this problem space Structure and Binding Evolutionary Aspects Structure and Binding 1. Structure identify stabilizing and targeting helices helical wheels, amphipathicity surface charge calculations 2. Binding generalized charge effects in approach geometric constraints specific charge-charge or hydrogen-bonding interactions ligand docking simulation enhanced with 3-D visualization and haptics Example of a student exercise: transcription regulator protein containing helix-loop-helix motif from Methanococcus maripaludis Interactive java helical wheel http://marqusee9.berkeley.edu/kael/helical.htm Evolutionary Aspects 1. Evolution over taxa of a particular HtH domain-containing protein tools: CLUSTALW, Phylip 2. Evolutionary utilization of this motif by different enzymes tools: BLAST, CLUSTALW, Domain search different sequence = same structure ? Tree for homeodomain engrailed from diverse organisms as analyzed through NCBI and Workbench BLASTP Homology search for Human “Engrailed Homolog 1” Homologous to mouse, chicken, rat, zebra fish, worm Human Engrailed Homolog 1 Conserved protein domain search using NCBI CD-Search