* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download No Slide Title - Ohio University
Oncogenomics wikipedia , lookup
Ridge (biology) wikipedia , lookup
Human genome wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression profiling wikipedia , lookup
Minimal genome wikipedia , lookup
Koinophilia wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Point mutation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Segmental Duplication on the Human Y Chromosome wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genomic imprinting wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Hybrid (biology) wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Genome evolution wikipedia , lookup
DiGeorge syndrome wikipedia , lookup
Y chromosome wikipedia , lookup
Microevolution wikipedia , lookup
Genome (book) wikipedia , lookup
Neocentromere wikipedia , lookup
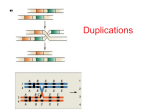
Chapter 17 Large-Scale Chromosomal Changes Changes in Chromosome Number Changes in Chromosome Shape Types of chromosome mutations all generated by natural mutagens—extreme temps, UV, chemicals, etc. Euploidy • euploidy: change of chromosome number involving 1 or more whole genomes – autopolyploidy = doubling of genome from “wild type” (e.g., tetraploid from diploid, hexaploid from triploid) – allopolyploid = doubling of genome from hybrid of two distinct taxa (e.g., varieties, species, genera) Examples of ploidy levels Balanced (normal meiosis) • 2n = diploid • 4n = tetraploid • 6n = hexaploid • 8n = octoploid • and so on Unbalanced (abnormal meiosis) • 1n = monoploid • 3n = triploid • 5n = pentaploid • 7n = heptaploid [usually hybrids of ploidy levels on left] Facts about polyploidy and allopolyploids • Uncommon in animals but abundant (ancient and ancestral?) in plants • Recent genetic research shows allopolyploids far more common than autopolyploids—different from theory • Many allopolyploids found with multiple origins—contrary to evolutionary paradigm of “single origin” for species Polyploids often produce larger structures, e.g., guard cells, pollen,... ...and fruits (e.g., tetraploid grapes) Meiotic pairing in triploids—> unbalanced gametes (sterility) Mules have 63 chromosomes, a mixture of the horse's 64 and the donkey's 62. The different structure and number prevents the chromosomes from pairing up properly and creating successful embryos, rendering mules infertile. Colchicine used to induce polyploidy A famous natural allohexaploid: Bread wheat (Triticum aestivum) Famous Examples of Allopolyploid Complexes • Appalachian Asplenium ferns—several diploids, triploid hybrids, several tetraploids • Domesticated coffee (Coffea arabica)-parentage documented through molecular cytogenetic “chromosome painting” • Dandelions, roses, blackberries--more complicated groups that also do agamospermy (sex without seeds) Evolutionary consequences of polyploidy • polyploids often more physiologically “fit” than diploids in extreme environments • polyploids reproductively isolated from original ploidy levels, may eventually differentiate • allopolyploids commonly occupy ecological niches not accessible to parental types • opportunities for gene silencing or chromosomal restructuring without disastrous consequences Monoploid plants grown in tissue culture Summary • polyploids common in plants • autoploids formed by doubling of “wild type” genome, allopolyploids from doubling of hybrid • allopolyploids far more common than autopolyploids • polyploids often more “fit” than parent(s), often in niches different from parent(s) • opportunities for evolutionary change through gene silencing or chromosome restructuring Facts about aneuploids • Rare in animals, always associated with developmental anomalies (if they survive) • Most well known examples in human genetic diseases • Common in plants, sometimes show phenotypes, sometimes not Extra chromosome 21 Down Syndrome Meiotic nondisjunction = aneuploid products Figure 16-12 step 1 Meiotic nondisjunction = aneuploid products Figure 16-12 step 2 Meiotic nondisjunction = aneuploid products Figure 16-12 step 3 Meiotic nondisjunction = aneuploid products Figure 16-12 step 4 Meiotic nondisjunction = aneuploid products Figure 16-12 step 5 Meiotic nondisjunction = aneuploid products Figure 16-12 step 6 Trisomics of Datura (jimsonweed) Large-Scale Chromosomal Changes Changes in Chromosome Structure Types of chromosome mutations Deletion loops in Drosophila genes missing from chromosome #2 #1 #2 Deletion loops in Drosophila Deletion origin of “cri du chat” syndrome see hear: http://www.youtube.com/watch?v=TYQrzFABQHQ Duplications following polyploidy in Saccharomyces Inversions cause diverse changes breakpoints between genes 1 breakpoint between genes, 1 within gene breakpoints within 2 genes Inversion loops at meiosis Paracentric inversions can lead to deletion products Paracentric inversions can lead to deletion products Paracentric inversions can lead to deletion products Paracentric inversions can lead to deletion products Paracentric inversions can lead to deletion products Paracentric inversions can lead to deletion products Pericentric inversions can lead to duplication-and-deletion products Figure 16-29 step 1 Pericentric inversions can lead to duplication-and-deletion products Figure 16-29 step 2 Pericentric inversions can lead to duplication-and-deletion products Figure 16-29 step 3 Pericentric inversions can lead to duplication-and-deletion products Figure 16-29 step 4 Reciprocal translocation revealed by molecular cytogenetics Chromosome segregation in reciprocal-translocation heterozygote Figure 16-30 step 1 Chromosome segregation in reciprocal-translocation heterozygote Figure 16-30 step 2 Chromosome segregation in reciprocal-translocation heterozygote Figure 16-30 step 3 Variegation resulting from gene’s proximity to heterochromatin Variegation in translocation heterozygote Chloroplast rearrangements • Great evolutionary significance in reconstructing relationships among land plant lineages • Can easily be screened for by PCR amplification of “universal” chloroplast gene primer pairs flanking large regions of chloroplast Judd et al. (2002) Chloroplast rearrangements • Major inversions found in certain groups of families of bryophytes, pteridophytes, gymnosperms and several groups of angiosperms • Loss of one copy of inverted repeat in a few families! • Numerous losses of certain introns across angiosperms (e.g., rpl2 in Cactaceae) • Differences in size of large single-copy region by expansion or contraction of intergenic spacers Summary • each different chromosomal change shows characteristic meiotic pairing as a “signature” • deletions in diploids often have grave consequences; in polyploids do not but may lead to differentiation of new organisms • duplications (in plants) generally have few or no consequences, often provide additional genes for evolutionary processes to act on (silencing, cooption by different functions)