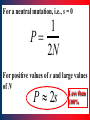* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download No Slide Title
Saethre–Chotzen syndrome wikipedia , lookup
Genome (book) wikipedia , lookup
Viral phylodynamics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Oncogenomics wikipedia , lookup
Gene expression programming wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Human genetic variation wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Koinophilia wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Frameshift mutation wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Point mutation wikipedia , lookup
Genetic drift wikipedia , lookup
Microevolution wikipedia , lookup
Gene Substitution Dan Graur 1 Gene substitution is the process whereby a mutant allele completely replaces the predominant or wild type allele in a population. Gene substitution occurs when a mutant allele arises in a population as a single copy in a single individual, increases its frequency to 1 (i.e., becomes fixed) after a certain number of generations. 2 Frequency of 1 Very low frequency 3 Not all mutants, however, reach fixation. In fact, the majority of them are lost after a few generations. 4 Very low frequency Frequency of 0 5 6 Fixation probability The probability that a particular allele will become fixed in a population depends on (1) its frequency (2) its selective advantage or disadvantage (3) the effective population size 7 The case of genic selection 1. three genotypes A1A1, A1A2, A2A2 2. fitness values: 1, 1 + s, 1 + 2s, The probability of fixation of A2 is: P 1 e 4Ne sq 4N s e 1 e where q is the frequency of allele A2. 8 P 1 e 4Ne sq 4N s e 1 e As s approaches 0 (neutral mutation), the equation reduces to P q The fixation probability for a neutral allele equals its frequency in the population. 9 A new mutant arising in a diploid population of size N has an initial frequency of 1/(2N). If the mutation is neutral the probability of fixation is P = 1/(2N). 10 For a neutral mutation, i.e., s = 0 1 P 2N For positive values of s and large values of N P 2s Less than 100% 11 Thus, if an advantageous mutation arises in a large population and its selective advantage over the rest of the alleles is small (up to ~5%), then the fixation probability is approximately twice its selective advantage. For example, the probability of fixation of a new codominant mutation with s = 0.01 is 2%. 12 13 Population size Neutral mutation Advantageous mutation (s = 0.01) Deleterious mutation (s = –0.001) 1,000 0.05% 2% 0.004% 10,000 0.005% 2% – 20 ~10 14 Mutation accumulation assay 15 Fixation Time The time required for the fixation (or the loss) of an allele depends on: (1) its frequency (2) its selective advantage or disadvantage (3) the effective population size 16 Conditional Fixation Time The time of fixation of mutants which do not undergo fixation is ∞. Thus, we only deal with the mean fixation time of those mutants that will eventually become fixed in the population. This variable is called the conditional fixation time. 17 Conditional Fixation Time In the case of a new neutral mutation whose initial frequency in a diploid population is by definition q = 1/(2N), the mean conditional fixation time is approximated by t 4N generations For a mutation with a selective advantage of s, the mean conditional fixation time is approximated by t (2 /s)ln( 2N) generations 18 Population Generation size time Neutral mutation 1,000,000 8 million years 2 years Advantageous Deleterious mutation mutation (s = 0.01) (s = –0.01) 5,800 years ? a. Less than 5,800 years b. More than 8 million years c. More than 5,800 but less than 8 million years d. 8 million years e. 5,800 years 19 Population Generation size time Neutral mutation 1,000,000 8 million years 2 years Advantageous Deleterious mutation mutation (s = 0.01) (s = –0.01) 5,800 years 5,800 years a. Less than 5,800 years b. More than 8 million years c. More than 5,800 but less than 8 million years d. 8 million years e. 5,800 years ✔ 20 Rate of Gene (or Allele) Substitution = number of mutants reaching fixation per unit time 21 Rate of Gene Substitution Neutral mutations: If neutral mutations occur at a rate of u per gene per generation, then the number of mutants arising at a locus in a diploid population of size N is 2Nu per generation. The probability of fixation for each neutral mutation is 1/(2N). The rate of substitution of neutral alleles is obtained by multiplying the total number of mutations by the probability of their fixation. 22 2Nu K 2N K u 23 24 Intuitive explanation: In a large population the number of mutations arising in every generation is high, but the fixation probability of each mutation is low. In a small population the number of mutations arising in every generation is low, but the fixation probability of each mutation is high. The rate of substitution for neutral mutations is independent of population size. 25 Rate of Gene Substitution Advantageous mutations: If advantageous mutations occur at a rate of u per gene per generation, then the number of mutants arising at a locus in a diploid population of size N is 2Nu per generation. The probability of fixation for each mutation is 2s. The rate of substitution of advantageous alleles is 4Nsu. 26 Deleterious mutations Neutral mutations Advantageous mutations Overdominant mutations 27 Mutational Meltdown: The double jeopardy of small populations It is possible for deleterious mutations to become fixed via genetic drift. Deleterious mutations occur more frequently than advantageous mutations. In small populations, random genetic drift is more important than selection. Small populations may be driven to extinction due to (1) accumulation of deleterious alleles, and (2) the fact that selection is too week to allow for advantageous mutations to accumulate. Michael Lynch 28 Multilocus models Previously, we assumed that the genetic transmission of an allele at one locus was independent of the transmission of another allele at a different locus. Under this assumption, we could treat each locus separately. In practice, however, the transmission of an allele at a locus may be dependent on the transmission of alleles at other loci. The most common cause for this lack of independence is linkage, i.e., the close physical proximity of two loci on the same chromosome and the finite rate of meiotic recombination in the sequence separating the two loci from each other. 29 Linkage equilibrium and disequilibrium A diploid organism. Two autosomal loci, A and B. Each locus with two alleles, A1 and A2 at locus A, and B1 and B2 at locus B. Linkage equilibrium occurs if the association between the alleles at the two loci is random. Linkage disequilibrium occurs if some combinations of alleles occur significantly more or significantly less frequently in a population than would be expected from a random association between the alleles at the two loci. 30 Hitchhiking and genetic draft A population withtwo neutral haplotypes, A2B1 and A2B2, coexist with frequencies of p2 and q2, respectively. An advantageous mutation, A1, arises on the haplotype carrying the B1 allele. (Completely arbitrary, it could have arisen on on the haplotype carrying the B2 allele.) Without the advantageous allele arising at locus A, the probability of fixation for alleles B1 and B2 would have been p2 and q2, respectively. The linkage to the advantageous allele A1, however, alters these expectations. On its way to fixation, the advantageous mutation A1 will carry along the linked B1 allele, and will ultimately render the population monomorphic at locus B. 31 Hitchhiking and genetic draft Advantageous mutations reduce or eliminate genetic variation at genetically linked sites (selective sweep). A neutral or even deleterious allele that is sufficiently tightly linked to a positively selected allele increases its frequency and may be swept to fixation (genetic hitchhiking). In genetic hitchhiking, only the initial conditions are stochastic, the rest of the process is deterministic (genetic draft). 32 Selective sweeps leave several characteristic molecular signatures in the population: 1.Eliminate nucleotide variation in the region of the genome close to the beneficial allele. 2.Cause an excess of high-frequency derived (new) alleles. 3.Create long-range associations with neighboring loci— the “long-range haplotype,” That is, a selective sweep will lead to creation of linkage disequilibrium over large swaths of DNA around the positively selected variant. 4.The positive selection in one population causes large frequency differences between populations—larger than for neutrally evolving alleles. 33 A selective sweep takes approximately 2N e 2ln generations. s In addition, the signature of positive selection may be identifiable for an additional amount of time, depending on the rates of mutation and recombination in the relevant region. 34 For how long after the fact can an evolutionary detective identify a selective sweep in the human population? 35 The estimated human effective population size is ~10,000. The mean generation time is 25 years. If a lucky mutation has a selective advantage of 5%, the sweep will be complete in ∼10,000 years. If a lucky mutation has a selective advantage of 1%, the sweep will be complete in ∼50,000 years. SELECTIVE SWEEPS CAN ONLY BE DETECTED FOR VERY SHORT PERIODS OF TIME 36 Detecting recent selective sweeps due to selection 37 Why are we (adult UH students) able to drink milk? 38 The digestion of the disaccharide lactose, the primary sugar present in milk, into its monosaccharide constituents, glucose and galactose, is catalyzed by a small-intestine enzyme called lactase-phlorizin hydrolase (LPH or lactase). 39 Lactase persistence In mammals, levels of lactase decline rapidly after weaning, and adults are not able to digest lactose. In humans, most individuals are unable to digest lactose as adults (lactose intolerant), i.e., they carry the trait lactase nonpersistence. Digestion of fresh milk in individuals who are lactose intolerant can result in diarrhea, which for most of human history was lethal. In populations in which the only source of milk is the mother, lactase nonpersistence is a selectively advantageous trait, since breastfeeding is a potent, albeit imperfect, contraceptive, which inhibits menstruation and delays resumption of ovulation. However, in some populations, a derived genetic trait has appeared, in which the ability to digest lactase is maintained in adults. Such individuals are lactose tolerant due to lactase persistence. This trait is particularly common in populations that have traditionally practiced dairying, i.e., in populations which can obtain milk extramaternally. Lactase persistence Lactase persistence arose at least twice in human populations The lactase-persistence haplotypes West Africa North Europe Bersaglieri et al. 2004 44 Background selection In the case of strong negative selection on a locus, genetically linked (neutral & advantageous) variants will also be removed, producing a decrease in the level of variation surrounding the locus under purifying selection. This process of purging non-deleterious alleles from the population due to spatial proximity to deleterious alleles is called background selection. Background selection is the opposite of Selective sweep. Because the deleterious mutations driving background selection are removed from the population, they are extremely difficult to detect. 45 Epistasis Previously, we assumed that each locus contributes independently to the fitness of the individual (i.e., different loci do not interact with one another in any manner that affects the fitness). Thus, each locus can be dealt with separately. This is not, however, always the case! Epistasis refers to interactions among alleles at different loci resulting in “non-independent effects.” In other words, epistasis occurs when the effects of an allele at one locus are modified by one or several alleles 46 at other loci. Epistasis Epistasis may be defined at the fitness level or at the level of the phenotype. We distinguish between functional epistasis, in which alleles at different loci produce non-independent phenotypic effects, and fitness epistasis, in which alleles at different loci nonindependently determine the fitness of their carrier, whether or not epistasis is detectable at the level of the phenotype. 47 Epistasis The genetic-background effect, according to which a mutation may have different effects on fitness depending on the genome in which it occurs, may be regarded as a generalized kind of fitness epistasis. 48 Epistasis Positive epistasis means that the phenotype (or the fitness) is higher than expected. Negative epistasis means that the phenotype (or the fitness) is lower than expected. In the literature, one may find different terms, such as, synergistic, diminishing, antagonistic, aggravating, ameliorating, buffering, compensatory, and reinforcing… Confusing! 49 Epistasis Positive epistasis means that the phenotype (or the fitness) is higher than expected. Negative epistasis means that the phenotype (or the fitness) is lower than expected. Mutation a at locus 1 increases IQ by 1 point. Mutation b at locus 2 increase IQ by 2 points. The two mutations together (say, following recombination) increase IQ by 12 points. Is the epistasis positive or negative? Is the epistasis functional or fitness epistasis? 50