* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Slide 1
Signal transduction wikipedia , lookup
List of types of proteins wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Transcription factor wikipedia , lookup
Gene expression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene regulatory network wikipedia , lookup
Promoter (genetics) wikipedia , lookup
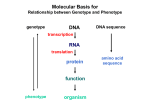
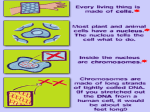
Gene Regulatory By Fatchiyah ([email protected]) Dept. of Biology, Brawijaya University 5/23/2017 fatchiyah, JB-UB Regulation of Genes What turns genes on (producing a protein) and off? Where (in which cells) is a gene turned on? When is a gene turned on or off? How many copies of the gene product are produced? 5/23/2017 fatchiyah, JB-UB Overview of Gene Control The mechanisms that control the expression of genes operate at many levels. source: Molecular Biology of the Cell (4th ed.), A. Johnson, et al. 5/23/2017 fatchiyah, JB-UB Multi-scale Model Problem Organizational Structure in Biology – Single Cells (Building blocks) – Tissues (organization of multiple cells) – Organism Fundamental Cellular Processes – – – – Inputs (Signal Transduction) Transcription Translation Outputs – adaptive processes 5/23/2017 fatchiyah, JB-UB DNA to chromosome M ovie: DNA to Chromosome The structure of DNA dictates the way it encodes genetic information. A strand of DNA consists of three types of fundamental chemical units, repeated many times. 5/23/2017 fatchiyah, JB-UB Prokaryote regulation Lac operon 5/23/2017 fatchiyah, JB-UB Transcriptional Regulation The transcription of each gene is controlled by a regulatory region of DNA relatively near the transcription start site (TSS). two types of fundamental components – short DNA regulatory elements – gene regulatory proteins that recognize and bind to them. 5/23/2017 fatchiyah, JB-UB Gene Signals in DNA CpG islands; methylation patterns; promotors Polymerase binding site; cofactors binding sites; enhancer; silencer transcription start sites; introns Donors; acceptors; branch point Termination signals 5/23/2017 fatchiyah, JB-UB Regulation of Genes (1) Transcription Factor (Protein) RNA polymerase (Protein) DNA ATG Regulatory Element 5/23/2017 Start site Gene Termination site fatchiyah, JB-UB source: M. Tompa, U. of Washington Regulation of Genes (2) Transcription ATG DNA Regulatory Element 5/23/2017 Gene fatchiyah, JB-UB source: M. Tompa, U. of Washington Regulation of Genes (3) ATG DNA mRNA source: M. Tompa, U. of Washington 5/23/2017 fatchiyah, JB-UB Protein Animation Transcription regulatory Post-Transcription regulation 5/23/2017 fatchiyah, JB-UB Post translation 5/23/2017 fatchiyah, JB-UB Cellular Processes 5/23/2017 fatchiyah, JB-UB Alignment of CpG island CpG island mRNA 5/23/2017 TATA-box Transcription start (homology-based assignment) fatchiyah, JB-UB Gene Regulatory Motif What is a motif? A subsequence (substring) that occurs in multiple sequences with a biological importance. Motifs can be totally constant or have variable elements. Protein Motifs often result from structural features. DNA Motifs (regulatory elements) – – – – 5/23/2017 Binding sites for proteins Short sequences (5-25) Up to 1000 bp (or farther) from gene Inexactly repeating patterns fatchiyah, JB-UB daf-19 Binding Sites in C. elegans -150 5/23/2017 source: Peter Swoboda GTTGTCATGGTGAC GTTTCCATGGAAAC GCTACCATGGCAAC GTTACCATAGTAAC GTTTCCATGGTAAC -1 che-2 daf-19 osm-1 osm-6 F02D8.3 fatchiyah, JB-UB Splice Motifs in A/T rich Organisms 5/23/2017 fatchiyah, JB-UB Motif Representing Consensus sequence: a single string with the most likely sequence(+/- wildcards) Regular expression: a string with wildcards, constrained selection Profile: a list of the letter frequencies at each position Sequence Logo: – graphical depiction of a profile – conservation of elements in a motif. 5/23/2017 fatchiyah, JB-UB Motif Logos: an Example 5/23/2017 fatchiyah, JB-UB (http://www-lmmb.ncifcrf.gov/~toms/sequencelogo.html) Finding Regulatory Motifs . . . Given a collection of genes with common expression, Find the (TF-binding) motif in common 5/23/2017 fatchiyah, JB-UB An Example: Hormonal Gene Regulation To understand the cellular mechanisms of hormonal (hydrophobic and Hydrophilic) gene regulation 5/23/2017 fatchiyah, JB-UB Hormonal Gene regulation 5/23/2017 OBJECTIVES: Describe basic mechanisms of gene regulation. Describe gene regulation at transcriptional level. Describe key elements present in the region of gene involved in gene transcription. Give a definition of Promoters, enhancers, Response elements, Transcription factors and co-activators. Aware that hydrophobic hormone receptor is a transcription factor. Describe basic organisation of steroid receptor molecule Describe how hydrophobic hormones regulate gene expression Describe how hydrophilic hormones regulate gene fatchiyah, JB-UB expression via cellular signalling molecules. Regulation at Transcriptional level Transcription by tissue specific factors: muscle cell specific transporter factor(myoD)- in myoblasts. Ker1=keratinocyte differentiating factor-skin cells. HNF-5=hepatic nuclear factor5- liver cells Hormone, growth factor or cellular messenger binding to response element: glucocorticoid response element (GRE): binds glucocorticoid receptor complex Alternative promoter: dystrophin gene 5/23/2017 fatchiyah, JB-UB Promoter Regions Many factor sites: c-fos, c-jun, c-myc/mac; Sp1, CREB, etc CREB C-myc/ max Sp-1 C/EBP RNA Pol II tgacgca cacgtg gggcgg ccatt -100 tata -35 Exon1 Exon2 Transcription ON mRNA TF II-B TF II-A TF II-D 5/23/2017 fatchiyah, JB-UB Protein Transcriptional regulation cis-acting transcriptional control sequences: Promoter, Response elements; Enhancers, silencer Transcription regulatory proteins (Transcription · factors) 5/23/2017 fatchiyah, JB-UB Cis acting transcriptional control sequences Promoters: Located in the immediate upstream region, serves to initiate transcription. Consists of combination of short sequence elements (ccaat box, tata box, gc box). Recognised by ubiquitous transcription factors. Promoters of gene that show tissue specific expression pattern often include cis-acting sequence elements that can be recognised by tissue specific transcription factors. 5/23/2017 fatchiyah, JB-UB Cis acting transcriptional control sequences Response elements: Found in selected genes whose expression is controlled by an external factor. Located short distance upstream of promoters. 5/23/2017 fatchiyah, JB-UB Cis acting transcriptional control sequences Enhancers: Positive regulatory elements, function independent of both orientation and distance from the genes they regulate. They contain elements recognised by ubiquitous transcription factors and tissue specific transcription factors. Silencers: negatively regulatory elements. 5/23/2017 fatchiyah, JB-UB Transcriptional regulatory proteins (Transcription factors) Transcription is regulated by a large number of DNA binding proteins. These DNA binding proteins mostly bind the response elements in a dimeric form, either as homodimers or or as heterodimers. The symmetry of these dimeric transcription factor is matched by their response elements which frequently posses an incomplete dyad symmetry. 5/23/2017 fatchiyah, JB-UB Structural motifs of the regulatory DNA binding protein Helix-turn-helix (HTH) motif: mediated DNA binding by fitting into major groove of the DNA helix, allowing precise alignment of the factor in relation to DNA sequence recognised Helix –loop helix: promotes both DNA binding and protein dimer formation. Zinc Finger: loops or fingers of amino acids that have a zinc ion at their core. The adjacent amino acid sequences form a α-helices that makes contact with DNA in the major groove Leucine Zipper: forms a dimer that grips the DNAdouble helix like a peg, by inserting into the major groove. 5/23/2017 fatchiyah, JB-UB DNA binding motifs in Proteins 5/23/2017 fatchiyah, JB-UB Properties of dimeric transcription factors. DNA binding module: Provides a non-selective affinity for DNA and the ability to recognise the specific base sequence of the response element. A dimerisation module allows protein to form an active dimeric state. Transcription activation (or inhibition) region: required for stimulation or inhibition of transcription 5/23/2017 fatchiyah, JB-UB Regulation of transcription factors Level of synthesis Covalent modification by phosphorylation /desphosphorylation Binding of small molecules can act as positive or negative allosteric effectors 5/23/2017 fatchiyah, JB-UB Specific transcription factors Tissue specific gene expression Cell growth and differentiation Response to lipid soluble hormones and second messenger molecules Cellular transcription factors required for the transcription of viral gene. 5/23/2017 fatchiyah, JB-UB Specific Hormonal Gene regulation Hormones regulate the function of their target cells by receptor mediated pathways. Water soluble hormones for which the lipid barrier of the plasma membrane is impenetrable, interact with their receptors at the cell surface Hydrophobic hormones that are lipid soluble and therefore can pass the cell membrane interact with another family of receptors that are intracellular (cytosolic or nuclear). These intracellular receptors function as hormone-regulated transcription factors, controlling the expression of specific target genes by interacting with regions known as hormone response elements located close to the promoter in the regulatory region 5/23/2017 fatchiyah, JB-UB Receptor-mediated induction of gene transcription by steroid hormones Steroid ligands Vacant Receptor hsp90 Pre-initiation complex RNA Pol-II + GTFs Coactivator protein(s) Activated receptors HRE 5/23/2017 TATA box fatchiyah, JB-UB Mechanism of gene activation by the steroid hormone receptors (transcription factors) The DNA binding domain is characterised by the presence of two zinc fingers. In the case of steroid hormones Zinc is coordinated by four cysteine residues. In other Zinc fingers Zn can be co-ordinated by two histidine and two cysteine residues. Although these are referred to as fingers, the tertiary structure is more like a fist made from beta sheet and α helical structure. DNA interacting residues are found in the α-helical part of the molecule. 5/23/2017 fatchiyah, JB-UB Mechanism of gene regulation by Hydrophilic hormones Intracellular second messenger cAMP and gene regulation. In the previous lecture you learnt that cAMP is produced as a result of activation of adenylate cyclase by G protein which in turn had been acivated by hormone which can not pass the hydrophobic cell membrane. This cAMP in turn acivates a cAMP depedent protein kinase (PKA) which modulates multiple aspects of cell function. PKA phosphorylates a transcription factor called CREB (cAMP response element binding protein). This is then translocated to the nucleus where it binds to a short pallinodromic sequence in the promoter region of cyclic AMP-regulated genes referred to as CRE. 5/23/2017 fatchiyah, JB-UB Thank You 5/23/2017 fatchiyah, JB-UB