* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download En/Spm-Mu
Deoxyribozyme wikipedia , lookup
Non-coding DNA wikipedia , lookup
Protein moonlighting wikipedia , lookup
RNA interference wikipedia , lookup
Genome evolution wikipedia , lookup
Gene desert wikipedia , lookup
Epitranscriptome wikipedia , lookup
Gene expression profiling wikipedia , lookup
List of types of proteins wikipedia , lookup
Community fingerprinting wikipedia , lookup
RNA silencing wikipedia , lookup
Non-coding RNA wikipedia , lookup
Expression vector wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene regulatory network wikipedia , lookup
Point mutation wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Molecular evolution wikipedia , lookup
Gene expression wikipedia , lookup
Bottromycin wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
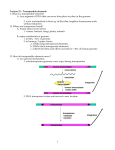
En/Spm transposable element system of maize Enhancer (En)/Suppressor-mutator (Spm) 13 bp perfect terminal inverted repeat P ATG ATG 8.3 kb tnpD tnpA Makes a 3-bp footprints (target site duplication) En/Spm Transposable elements cis Determinants for Excision: Approx. 180 bp at the 5’ end and 300 bp of 3’ end represent cis determinants. Contained in these regions are reiterations of a 12-bp sequence motif that is recognized by TNPA protein with 6 motifs present at 5’ end and 8 at 3’ end. Trans-factors: TNPA and TNPD (transposase complex) TNPA brings the two ends together and causes DNA bending, TNPD cleaves TNPA TNPD The “suppressor” function of En/Spm Transposable elements En/Spm have a unique feature: they act as suppressor of gene function The non-autonomous derivative of En/Spm (dSpm) when inserted into a gene causes reduced gene expression of that gene instead of knocking it out. The residual gene activity is due to the spicing of dSpm from pre-mRNA. However, if trans-factors TNPA is present then gene activity is knocked out i.e. pre-mRNA is not formed. TNPA binding with dSpm probably causes steric hindrance for RNA polymerase. Mutator Transposable element system of maize Mutator (Mu) trait was first identified by Robertson (1978) as a heritable high forward-mutation rate exhibited by maize lines. Many of these de novo mutations exhibited somatic instability, primarily apparent reversion to wildtype. This phenomenon was found to be associated with Mu transposable elements. Autonomous MuDR element (4942-bp): 200-bp TIR, create 9-bp target site duplication. TIR P 2.8-kb RNA (823 aa peptide) 1-kb RNA (207 aa peptide) Both proteins tightly bind to TIRs P TIR Several Mu elements (subfamilies) exist that contain variable internal sequence. These are non-autonomous derivatives of MuDR. Applications: 1. Mu elements are known to transpose to any locus, especially genes, therefore it is very useful for creating tagged mutations. 2. Mutator’s frequent transposition activity (even to unlinked locus) is reminiscent of P element system of Drosophila. In Drosophila, P elements have been used as vectors to increase the efficiency of transgene integration in the injected oocytes. Therefore, Mu is an attractive system for increasing transformation efficiency of maize. The strategy would involve introducing gene-of-interest in a Mu element and then injecting that into maize cells containing MuDR (MuDR based cloning in impossible because MuDR is unstable in E. coli). However, no report has so far been published that describes the success for this strategy.