* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download lect21
Nucleic acid analogue wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Ligand binding assay wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Peptide synthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Proteolysis wikipedia , lookup
Catalytic triad wikipedia , lookup
Protein structure prediction wikipedia , lookup
Metalloprotein wikipedia , lookup
Genetic code wikipedia , lookup
Epitranscriptome wikipedia , lookup
Biochemistry wikipedia , lookup
Amino acid synthesis wikipedia , lookup
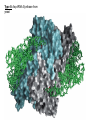
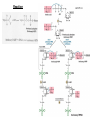
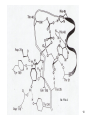
Lecture #21: Aminoacyl tRNA Synthetases: The Ancient Enzyme (E.C.# 6.1.1.1) -RS are ancient enzymes over 3.5 billion years old -evolution/development closely connected with genetic code -RS enzymes are at the centre of research on the origin of life Reaction: Amino acid + tRNA + ATP aminoacyl—tRNA + AMP + PPi Classification: -at least one aminoacyl tRNA synthetase for each amino acid -grouped into two classes: -Class I and II -each class divided into three subclasses a, b, and c -each class originated from a single domain ancestor De Pouplana & 1 Schimmel 2001 TiBS 26, 591-596. Class I -11 enzymes with active site that has Rossmann fold (parallel -sheet domain) -subclass Ia -hydrophobic amino acids (Ile, Leu, Val) -sulfur amino acids (Met and Cys) -Arg -subclass Ib -charged amino acids (Glu, Lys) and derivative Gln -subclass Ic -aromatic amino acids (Tyr and Trp) -acylate the 2’-hydroxyl group of terminal adenosine of tRNA Class II -10 members that possess a 7-stranded -sheet with flanking -helices -subclass IIa -aliphatics (Ala, Pro) -polar (Ser, Thr, His) -Gly -subclass IIb -charged amino acids (Asp, Lys) and derivative Asn -subclass IIc -aromatic amino acid (Phe) -acylate the 3’-hydroxyl group of terminal adenosine of tRNA 2 Type I: Gln-tRNA Synthase from E. coli 3 Type II: Asp-tRNA Synthase from yeast 4 -wide variety of structural types, especially in eukaryotes -, 2, 4, and 22 with total Mr values ranging from 50,000 – 300,000 -subunit Mr values are larger in eukaryotes and involves an N-terminal extension (50 – 300 amino acids) V&V:T32-4 5 Reaction L4:F27-14 6 Tyrosyl tRNA Synthetase -X-ray crystallographic and protein engineering studies have provided insight into the catalytic mechanism of tyrosyl-tRNA synthase, a Class I dimer of 47 kDa subunits -in centre of each subunit is a 6-stranded -sheet structure with 5 longer helices and several shorter ones -a number of complexes have been examined by x-ray crystallography involving AMP, ATP, tyrosine, and tyrosyl-AMP (highly stable) -amino terminal 320 residues are needed for the activation reaction -carboxy terminal 99 residues participate in the binding of tRNA and the formation of the tyrosyl-tRNA -activated intermediate is stable in the absence of matching tRNA and is bound to enzyme with 12 H-bonds Tyr-tRNA synthetase with tyrosineadenylate bound in active site 7 Overview Chains Residues Mol. Weight [D] Chain Type 3TS1:_ 419 47290 Protein Download all chains in FASTA format Secondary Structure Elements given below are documented in the Help Section Chain 3TS1:_ Tyrosyl-Transfer RNA Synthetase (E.C. 6.1.1.1) Complexed With Tyrosinyl Adenylate - Chain _ Type Protein Molecular Weight 47290 Number of 419 Residues Number of Alpha 17 Content of Alpha 37.95 Number of Beta 6 Content of Beta 6.21 Compound Sequence and secondary structure 1 MDLLAELQWR GLVNQTTDED GLRKLLNEER VTLYCGFDPT ADSLHIGHLA HHHHHHHH T SEES HH HHHHHHHHS EEEEEE S SSS BTTTHH 51 TILTMRRFQQ AGHRPIALVG GATGLIGDPS GKKSERTLNA KETVEAWSAR HHHHHHHHHH TT EEEEEE TTTTTT T T SS HHHHHHHHHH 101 IKEQLGRFLD FEADGNPAKI KNNYDWIGPL DVITFLRDVG KHFSVNYMMA HHHHHHHHS SS SSS EE EETHHHHTT HHHHHHHTG GGTTHHHHTT 151 KESVQSRIET GISFTEFSYM MLQAYDFLRL YETEGCRLQI GGSDQWGNIT SHHHHTTTTT HHHHTHH HHHHHHHHHH HHHH EEE E GGGHHHHH 201 AGLELIRKTK GEARAFGLTI PLVTKADGTK FGKTESGTIW LDKEKTSPYE HHHHHHHHHH EEEE SSSS TT SS B SSTTTTTHHH 251 FYQFWINTDD RDVIRYLKYF TFLSKEEIEA LEQELREAPE KRAAQKTLAE HHHHHHTTTH HHHTHHHHHH HHHHHH HHHHHHHTTT TTHHHHHHHH 301 EVTKLVHGEE ALRQAIRISE ALFSGDIANL TAAEIEQGFK DVPSFVHEGG HHHHHHHTHH HHHHHHHH 351 DVPLVELLVS AGISPSKRQA REDIQNGAIY VNGERLQDVG AILTAEHRLE 401 GRFTVIRRGK KKYYLIRYA 8 H-bonds and Specificity -side chains implicated in H-bonds have systematically been replaced by non-Hbonding residues (eg., Cys 35 Gly, Tyr 34 Phe) -side chains responsible for specificity of the enzyme for tyrosine as opposed to phenylalanine are: Tyr 34 and Asp 176 -in ribose binding site: Cys 35, Thr 51, and His 48 -Cys 35 is conserved in bacterial tyrosyl-tRNA synthases but replacement resulted in an enzyme with 30% of wild-type activity -results of mutagenesis showed that the different types of H-bonds made different contributions to the binding energy -mutation of an uncharged side chain (Tyr 169) that forms a hydrogen bond to a charged group on the substrate (the -amino group) weakens the binding by 15.5 kJ/mol -mutation of a side chain (Tyr 34) that forms an H-bond to an uncharged group (the phenolic OH group of tyrosyl-AMP) weakens the binding by only 2.2 kJ/mol -Thr 51 forms an unfavorable H-bond with the ribose of tyrosyl AMP; it could form a stronger H-bond with water promoting the dissociation of the tyrosyl-AMP complex -mutation of Thr 51 to Pro or Ala improved the kcat by 50- and 2-fold, respectively 9 S4: F34-9 10 G3:F30-7a 11 Catalysis -reaction proceeds by an in-line displacement mechanism where the tyrosyl carboxylate is the attacking nucleophile and the pyrophosphate is the leaving group - phosphorus atom in the transition state is pentavalent and the geometry of this state is trigonal bipyramidal (cf. RNAse A) -model for transition state includes the H-bonding of the -phosphate group to the side chains of Thr 40 and His 45 -double mutant, T40A and H45A, has a decreased kcat of 3.6 x 106 fold but binding affinity of the enzymes for ATP and tyrosine were unaltered shows Thr 40 and His 45 important for catalysis but not substrate binding -these residues likely interact with the phosphate group in the transition state but not in the initial enzyme-substrate complex -selective binding believed to be triggered by the large shift in position of the pyrophosphate unit accompanying the tetrahedral to bipyramidal geometry change -classic instance that “the essence of catalysis is the selective stabilization of the transition state” 12 What are the catalytic residues? -perhaps there are none because the carboxylate group of Tyr is an intrinsically effective nucleophile, ATP is already activated, and Mg2+--PPi is a good leaving group -enzyme may simply accelerate the reaction by a factor of 4 x 104 by bringing Tyr and ATP together and it may gain another factor of 3 x 105 mainly by binding phosphate in the transition state -since ATP, amino acid, and pyrophosphate can each bind to the enzyme separately, the reaction is randomorder ternary type -in most cases the rate of the first reaction is 10 – 100 times the rate of the second reactions, but in some enzymes the rates are nearly equal 13 S4:F34-10 Editing/Proofreading by Aminoacyl tRNA Synthetases -aminoacyl tRNA synthetases are highly selective in their recognition of both the amino acid to be activated and the prospective tRNA acceptor -tRNA molecules that accept different amino acids have different base sequences so they can readily be distinguished by their synthetases How do these enzymes discriminate between Ile and Val? -extra methylene group in Ile provides additional binding energy of –12 kJ/mol which favours the activation of Ile by isoleucine tRNA synthetase by a factor of 200 -however, concentration of Val in vivo is 5 times that of Ile Val should mistakenly be incorporated 1 in 40 times -observed frequency is 1 in 3000 times editing function -mistakenly activated Val is not transferred to tRNA specific for Ile -this tRNA promotes the hydrolysis of Val—AMP and prevents the erroneous incorporation into proteins -this hydrolysis frees the synthetase to activate and transfer Ile, the correct amino acid -the enzyme avoids hydrolyzing the Ile-AMP because the hydrolytic site is just large enough to accommodate Val—AMP but too small to allow the entry of IleAMP 14 S4:F34-12 15 What about amino acids that are nearly identical in size (Val and Thr)? -the aminoacyl tRNA synthetase for Val contains two adjacent catalytic sites, one for the acylation of tRNA and the other for the hydrolysis of incorrectly acylated tRNA -Val is preferred over Thr in the acylation reaction because the acylation site is more hydrophobic -threonyl tRNA is hydrolyzed more rapidly because the hydrolysis site is more hydrophilic -the synthetase for Val does most of the editing at the level of the aminoacyltRNA whereas the one for Ile does so at the level of the aminoacyl-AMP -most aminoacyl tRNA synthetases contain hydrolytic sites in addition to acylation sites -complementary pairs of sites function as a double sieve to assure high fidelity -the acylation site rejects amino acids that are larger than the correct one whereas the hydrolytic site destroys activated intermediates that are smaller than the correct species -hydrolytic proofreading is essential to the fidelity of many aminoacyl tRNA synthetases -some synthetases do not require editing functions because the binding of other amino acids is much weaker eg., tyrosyl-tRNA synthetases bind Tyr 104 x stronger than Phe 16 Aminoacyl Synthetase Recognition of tRNA -some synthetases recognize their tRNA partner based on the anticodon tRNAAla is recognized at the 3:70 position in the 3’ acceptor stem of this 76-nucleotide molecule -tRNACys differs from tRNAAla at 40 positions and contains a C-G basepair at the 3:70 position -when the C-G basepair is changed to G-U at the 3:70 position of the tRNACys then alanyl-tRNA synthetase recognizes it as though it were tRNAAla -a microhelix containing 24 of the 76 nucleotides of the native tRNA is specifically recognized by alanyl-tRNA synthetase G3:F30-8 17