* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Three main topics for this Intro lecture
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Paracrine signalling wikipedia , lookup
Genetic code wikipedia , lookup
Signal transduction wikipedia , lookup
Gene expression wikipedia , lookup
Point mutation wikipedia , lookup
Expression vector wikipedia , lookup
Biochemistry wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Magnesium transporter wikipedia , lookup
Metalloprotein wikipedia , lookup
Interactome wikipedia , lookup
Structural alignment wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Anthrax toxin wikipedia , lookup
Simple Rearrangements
Reversals
1
2
3
9
8
4
7
1, 2, 3, 4, 5, 6, 7, 8, 9, 10
•
Blocks represent conserved genes.
6
5
10
1
2
Reversals
3
9
8
4
7
1, 2, 3, -8, -7, -6, -5, -4, 9, 10
10
6
5
Blocks represent conserved genes.
In the course of evolution or in a clinical context, blocks 1,…,10
could be misread as 1, 2, 3, -8, -7, -6, -5, -4, 9, 10.
Types of Rearrangements
Reversal
1 2 3 4 5 6
1 2 -5 -4 -3 6
Translocation
1 2 3
45 6
1 26
4 53
Fusion
1 2 3 4
5 6
1 2 3 4 5 6
Fission
Sorting by reversals: 5 steps
Step
Step
Step
Step
Step
Step
0: p 2 -4
1:
2 3
2:
2 3
3:
2 3
4:
-8 -7
5: g 1 2
-3
4
4
4
-6
3
5
5
5
5
-5
4
-8
-8
6
6
-4
5
-7
-7
7
7
-3
6
-6
-6
8
8
-2
7
1
1
1
-1
-1
8
Sorting by reversals: 4 steps
Step
Step
Step
Step
Step
0: p 2 -4 -3
1:
2 3 4
2:
-5 -4 -3
3:
-5 -4 -3
4: g 1 2 3
5
5
-2
-2
4
-8
-8
-8
-1
5
-7
-7
-7
6
6
-6
-6
-6
7
7
1
1
1
8
8
Sorting by reversals: 4 steps
Step
Step
Step
Step
Step
0: p 2 -4 -3
1:
2 3 4
2:
-5 -4 -3
3:
-5 -4 -3
4: g 1 2 3
5
5
-2
-2
4
-8
-8
-8
-1
5
-7
-7
-7
6
6
-6
-6
-6
7
7
1
1
1
8
8
What is the reversal distance for this
permutation? Can it be sorted in 3 steps?
From Signed to Unsigned Permutation (Continued)
• Construct the breakpoint graph as usual
• Notice the alternating cycles in the graph between every other vertex
pair
• Since these cycles came from the same signed vertex, we will not be
performing any reversal on both pairs at the same time; therefore, these
cycles can be removed from the graph
0
5
6 10 9 15 16 12 11 7 8 14 13 17 18 3
4
1 2 19 20 22 21 23
Reversal Distance with Hurdles
• Hurdles are obstacles in the genome rearrangement problem
• They cause a higher number of required reversals for a permutation
to transform into the identity permutation
• Let h(π) be the number of hurdles in permutation π
• Taking into account of hurdles, the following formula gives a
tighter bound on reversal distance:
d(π) ≥ n+1 – c(π) + h(π)
Median Problem
Goal: find M so that DAM+DBM+DCM is minimized
NP hard for most metric distances
Genome Enumeration for
Multichromosome Genomes
.
.
.
Genome Enumeration
For genomes on gene {1,2,3}
2
.
.
.
-3
1
-1
$
23
.
.
.
2
.
.
.
3
$
‹ 1, 2, 3 ›
-3
$
‹ 1, 2, -3 ›
3
$
‹ 1, 2 › ‹ 3 ›
-3
$
‹ 1, 2 › ‹ -3 ›
...
...
-2-3
...
3
...
-3
...
Rearrangement Phylogeny
Compute A Given Tree (Start)
Compute A Given Tree (First Median)
Compute A Given Tree (Second Median)
Compute A Given Tree (Third Median)
Compute A Given Tree (After 1st Iteration)
Binary Encoding
MLBE Sequences
Experimental Results (Equal Content)
80% inversion, 20% transposition
An Example—New Genomes
1 2
1 -4
…
3
5
1 3 5 7 9
1 5 9 -7 3
…
4
2
5 6
8 10
7 8 9 10
9 -7 -6 3
Jackknifing Rate
Support Value Threshold - FP
Up to 90% FP can be identified with 85% as the
threshold
Jackknife Properties
• Jackknifing is necessary and useful for gene
order phylogeny, and a large number of
errors can be identified
• 40% jackknifing rate is reasonable
• 85% is a conservative threshold, 75% can
also be used
• Low support branches should be examined
in detail
Protein
In-silico Biochemistry
• Online servers exist to determine many
properties of your protein sequences
• Molecular weight
• Extinction coefficients
• Half-life
• It is also possible to simulate protease digestion
• All these analysis programs are available on
• www.expasy.ch
Analyzing Local Properties
• Many local properties are important for the function of
your protein
• Hydrophobic regions are potential transmembrane domains
• Coiled-coiled regions are potential protein-interaction
domains
• Hydrophilic stretches are potential loops
• You can discover these regions
• Using sliding-widow techniques (easy)
• Using prediction methods such as hidden Markov Models
(more sophisticated)
Sliding-window Techniques
• Ideal for identifying strong
signals
• Very simple methods
• Few artifacts
• Not very sensitive
• Use ProtScale on
www.expasy.org
• Make the window the same
size as the feature you’re
looking for
www.expasy.org/cgi-bin/protscale.pl
www.expasy.org/cgi-bin/protscale.pl
www.expasy.org/cgi-bin/protscale.pl
www.expasy.org/cgi-bin/protscale.pl
Hphob. / Eisenberg
Transmembrane Domains
• Discovering a transmembrane
domain tells you a lot about your
protein
• Many important receptors have 7
transmembrane domains
• Transmembrane segments can be
found using ProtScale
• The most accurate predictions
come from using TMHMM
Using TMHMM
• TMHMM is the best method for predicting transmembrane
domains
• TMHMM uses an HMM
• Its principle is very different from that of ProtScale
• TMHMM output is a prediction
TMHMM vs. ProtScale
>sp|P78588|FREL_CANAX Probable ferric reductase transmembrane component OS=Candida albicans
GN=CFL1 PE=3 SV=1
MTESKFHAKYDKIQAEFKTNGTEYAKMTTKSSSGSKTSTSASKSSKSTGSSNASKSSTNA
HGSNSSTSSTSSSSSKSGKGNSGTSTTETITTPLLIDYKKFTPYKDAYQMSNNNFNLSIN
YGSGLLGYWAGILAIAIFANMIKKMFPSLTNNLSGSISNLFRKHLFLPATFRKKKAQEFS
IGVYGFFDGLIPTRLETIIVVIFVVLTGLFSALHIHHVKDNPQYATKNAELGHLIADRTG
ILGTFLIPLLILFGGRNNFLQWLTGWDFATFIMYHRWISRVDVLLIIVHAITFSVSDKAT
GKYKNRMKRDFMIWGTVSTICGGFILFQAMLFFRRKCYEVFFLIHIVLVVFFVVGGYYHL
ESQGYGDFMWAAIAVWAFDRVVRLGRIFFFGARKATVSIKGDDTLKIEVPKPKYWKSVAG
GHAFIHFLKPTLFLQSHPFTFTTTESNDKIVLYAKIKNGITSNIAKYLSPLPGNTATIRV
LVEGPYGEPSSAGRNCKNVVFVAGGNGIPGIYSECVDLAKKSKNQSIKLIWIIRHWKSLS
WFTEELEYLKKTNVQSTIYVTQPQDCSGLECFEHDVSFEKKSDEKDSVESSQYSLISNIK
QGLSHVEFIEGRPDISTQVEQEVKQADGAIGFVTCGHPAMVDELRFAVTQNLNVSKHRVE
YHEQLQTWA
Search with Accession number P78588
http://www.uniprot.org/uniprot/
www.cbs.dtu.dk/services/TMHMM-2.0
www.cbs.dtu.dk/services/TMHMM-2.0
Predicting Post-translational
Modifications
• Post-translational modifications often occur on similar motifs in
different proteins
• PROSITE is a database containing a list of known motifs, each
associated with a function or a post-translational modification
• You can search PROSITE by looking for each motif it contains in
your protein (the server does that for you!)
• PROSITE entries come with an extensive documentation on each
function of the motif
Searching for
PROSITE Patterns
• Search your protein against PROSITE on ExPAsy
• www.expasy.org/tools/scanprosite
• PROSITE motifs are written as patterns
• Short patterns are not very informative by themselves
• They only indicate a possibility
• Combine them with other information to draw a conclusion
• Remember: Not everything is in PROSITE !
www.expasy.org/tools/scanprosite
P12259
www.expasy.org/tools/scanprosite
Interpreting PROSITE Patterns
• Check the pattern function: Is it compatible with the protein?
• Sometimes patterns suggest nonexistent protein features
• For instance : If you find a myristoylation pattern in a prokaryote,
ignore it; prokaryotic proteins have no myristoylation !
• Short patterns are more informative if they are conserved across
homologous sequences
• In that case, you can build a multiple-sequence alignment
• This slide shows an example
Patterns and Domains
• Patterns are usually the most striking feature of the
more general motifs (called domains)
• Domains are less conserved than patterns but usually
longer
• In proteins, domain analysis is gradually replacing
pattern analysis
Protein Domains
• Proteins are usually
made of domains
• A domain is an
autonomous folding
unit
• Domains are more than
50 amino acids long
• It’s common to find
these together:
• A regulatory domain
• A binding domain
• A catalytic domain
Discovering Domains
• Researchers discover domains by
• Comparing proteins that have similar functions
• Aligning those proteins
• Identifying conserved segments
• A domain is a multiple-sequence alignment
formulated as a profile
• For each column, a domain indicates which amino
acid is more likely to occur
Domain Collections
• Scientists have been discovering and characterizing protein
domains for more than 20 years
• 8 collections of domains have been established
• Manual collections are very precise but small
• Automatic collections are very extensive but less informative
• These collections
• Overlap
• Have been assembled by different scientists
• Have different strengths and weaknesses
• We recommend using them all!
The Magnificent 8
• Pfam is the most extensive manual collection
• Pfam is often used as a reference
Searching Domain Collections
• Domains in Pfam often include known functions
• A match between your protein and a domain is desirable
• A match is a potential indication of a function
• This is VERY informative for further research!
• Three servers exist to compare proteins and domain
collections:
• InterProScan
www.ebi.ac.uk/interproscan
• CD-Search (conserved Domain)
www.ncbi.nih.nlm.gov
• Motif Scan
www.ch.embnet.org
Using InterProScan
• InterProScan is the most
comprehensive search engine
for domain databases
• Makes it possible to compare
alternative results on most
collections
• Does not provide a statistical
score
>sp|P53539|FOSB_HUMAN Protein fosB OS=Homo sapiens GN=FOSB PE=1 SV=1
MFQAFPGDYDSGSRCSSSPSAESQYLSSVDSFGSPPTAAASQECAGLGEMPGSFVPTVTA
ITTSQDLQWLVQPTLISSMAQSQGQPLASQPPVVDPYDMPGTSYSTPGMSGYSSGGASGS
GGPSTSGTTSGPGPARPARARPRRPREETLTPEEEEKRRVRRERNKLAAAKCRNRRRELT
DRLQAETDQLEEEKAELESEIAELQKEKERLEFVLVAHKPGCKIPYEEGPGPGPLAEVRD
LPGSAPAKEDGFSWLLPPPPPPPLPFQTSQDAPPNLTASLFTHSEVQVLGDPFPVVNPSY
TSSFVLTCPEVSAFAGAQRTSGSDQPSDPLNSPSLLAL
www.ebi.ac.uk/InterProScan
www.ebi.ac.uk/InterProScan
The CD-Search Output
• CD search is less extensive than that of InterProScan
• Results come with a a statistical evaluation (E-value)
• 10e-15
Low E-value
Good match
• 2.1
High E-value
Bad match
www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
Predicting Functions
with Domains
• Finding a match with a domain having a catalytic function is
good news . . . but what, exactly, does it mean?
• A match indicates that your sequence has the domain
structure . . . but does it also have the function?
• You cannot say before looking into these details:
• Where are the catalytic residues on the domain?
• Does your sequence have the right residues at these positions?
Looking into the Details
• Catalytic residues are normally highly conserved in
domains
• Motif Scan makes it possible to check whether these
important residues are conserved in your sequence
• High bar above 0 = Highly conserved residues
• Green = Your sequence has an expected residue
• Red = Your sequence has an unexpected residue
Looking into the Details (cont’d.)
R (Arginine) is highly
expected at this position
High bar
Potential active site
If your protein has an arginine
on this position . . .
Bar is filled with green
Your protein could be active
myhits.isb-sib.ch/cgi-bin/motif_scan
Protein 3D Structure
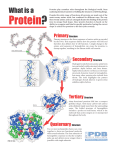
Primary, Secondary
and Tertiary Structures
• Proteins are made of 20 amino acids
• Proteins are on average 400 amino acids
long
• Protein structure has 3 levels:
• The primary structure is the sequence of a
protein
• The secondary structure is the local structure
• The tertiary structure is the exact position of
each atom on a 3D model
Secondary Structures
• Helix
• Amino acid that twists like a spring
• Beta strand or extended
• Amino acid forms a line without
twisting
• Random coils
• Amino acid with a structure
neither helical nor extended
• Amino-acid loops are usually coils
Guessing the Secondary Structure
of Your Protein
• Secondary structure predictions are good
• If your protein has enough homologues, expect
80% accuracy
• The most accurate secondary structure prediction
server is PSIPRED
PSIPRED Output
• Conf = Confidence
• 9 is the best, 0 the worst
• Pred = Every amino acid is assigned a letter:
• C for coils
• E for extended or beta-strand
• H for helix
>gi|15892329|ref|NP_360043.1| translocation protein TolB [Rickettsia conorii str. Malish 7]
MRNIIYFILSLLFSVTSYALETINIEHGRADPTPIAVNKFDADNSAADVLGHDMVKVISNDLKLSGLFRP
ISAASFIEEKTGIEYKPLFAAWRQINASLLVNGEVKKLESGKFKVSFILWDTLLEKQLAGEMLEVPKNLW
RRAAHKIADKIYEKITGDAGYFDTKIVYVSESSSLPKIKRIALMDYDGANNKYLTNGKSLVLTPRFARSA
DKIFYVSYATKRRVLVYEKDLKTGKESVVGDFPGISFAPRFSPDGRKAVMSIAKNGSTHIYEIDLATKQL
HKLTDGFGINTSPSYSPDGKKIVYNSDRNGVPQLYIMNSDGSDVQRISFGGGSYAAPSWSPRGDYIAFTK
ITKGDGGKTFNIGIMKACPQDDENSERIITSGYLVESPCWSPNGRVIMFAKGWPSSAKAPGKNKIFAIDL
TGHNEREIMTPADASDPEWSGVLN
bioinf.cs.ucl.ac.uk/psipred//?program=psipred
bioinf.cs.ucl.ac.uk/psipred//?program=psipred
bioinf.cs.ucl.ac.uk/psipred//?program=psipred
bioinf.cs.ucl.ac.uk/psipred//?program=psipred
Predicting Other
Secondary Features
• It is also possible to predict these accurately:
•
•
•
•
Transmembrane segments
Solvent accessibility
Globularity
Coiled/coil regions
• All these predictions have an expected accuracy
higher than 70%
Servers
•
•
•
•
www.predictprotein.org
cubic.bioc.columbia.edu/predictprotein
www.sdsc.edu/predicprotein
www.cbi.pku.edu.cn/predictprotein
Predicting 3D Structures
• Predicting 3D structures from sequences only is almost impossible
• The only reliable way to establish the 3D structure of a protein is to
make a real-world experiment in
• X-ray crystallography
• Nuclear magnetic resonance (NMR)
• Structures established this way are conserved in the PDB database
• “The PDB of my protein” is synonymous with “The structure of my
protein”
Retrieving Protein Structures
from PDB
• All PDB entries are 4-letter words!
• 1CRZ, 2BHL . . .
• Sometimes the chain number is added:
• 1CRZA, 1CRZB . . .
• To access all PDB entries, go to www.rcsb.org
• PDB contains 42,000 entries
• PDB contains the structure of 16,000 unique proteins or RNAs
• You can download the coordinates and display the structure
www.rcsb.org
www.rcsb.org
Displaying a PDB Structure
• You can use any of the online
viewers to display the structure
• They will let you rotate the
structure, zoom in and out, or
color it
• PDB files themselves are not
human-readable
Predicting the Structure
of Your Protein
• The bad news:
• It is very hard to predict protein 3D structures
• The good news:
• Similar proteins have similar structures
• If your favorite protein has a homologue with a known structure .
..
• You can do homology modeling
• How?
• Start with a BLAST (more about that in the next slide)
ncbi.nlm.nih.gov/BLAST
ncbi.nlm.nih.gov/BLAST
BLASTing PDB for Structures
• BLAST your protein against
PDB
• If you get a very good hit, it
means PDB contains a
protein similar to yours
• Your protein and this hit
probably have the same
structure
Be Careful!
• Sometimes only one of the domains contained in your protein
has been characterized
• If that’s the case, the PDB will only contain this domain
• Always check the alignments
• Red line = full protein in PDB
• Blue line = one domain only in this entry
Structures and Sequences
• Highly conserved sequences are often important in the structure
• Make a multiple-sequence alignment to identify these important
positions
• Highly conserved positions are either in the core or important for
protein/protein interactions
3D Predictions
• If you want to predict the structure of your protein
automatically, try the Swiss Model
• Swiss Model makes the BLAST for you
• The program does a bit of homology modeling
• The process delivers a new PDB entry
• You can access it at swissmodel.expasy.org
• Swiss Model gives good results for proteins having
homologues in PDB
zhanglab.ccmb.med.umich.edu/I-TASSER/
zhanglab.ccmb.med.umich.edu/I-TASSER/
3D-BLAST
• Use this technique if you have a structure and you
want to find other similar structures
• Use VAST or DALI to look for proteins having the
same 3D shape as yours
• www.eb.ac.uk/dali
• www.ncbi.nlm.nih/vast
3D Movements
• Most proteins need to move to do their job
• Predicting protein movement is possible using
molecular dynamics
• Check out this site: molmolvdb.mbb.yale.edu
• Good molecular dynamics requires extremely powerful
computers
• Don’t expect miracles from standard online resources