* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Bionano
Epitranscriptome wikipedia , lookup
Expanded genetic code wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Protein adsorption wikipedia , lookup
Polyadenylation wikipedia , lookup
Genetic code wikipedia , lookup
Molecular cloning wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Non-coding RNA wikipedia , lookup
Non-coding DNA wikipedia , lookup
Point mutation wikipedia , lookup
List of types of proteins wikipedia , lookup
Molecular evolution wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Two-hybrid screening wikipedia , lookup
DNA supercoil wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Gene expression wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
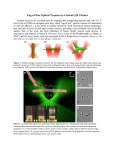
Bionano Part 1. Biocatalytic Synthesis of Polymers with Precisely Defined Structures, Deming, Conticello & Tirrell, in Nanotechnology, G. Timp, ed., Springer-Verlag, New York, 1999. Part 2. Measuring molecular machines (Stephen Block website). Part 3 (yet to be completed) Mixing bio building blocks with synthetic ones (Seth Fraden) Part 1 In Nanotechnology, ed. Gregory L. Timp, Springer-Verlag, New York, 1999 Uniform polymers Dendrimer Merrifield: good for ~60 residues, no proofreading Templated synthesis, as in protein biosynthesis. Details of protein biosynthesis Getting E. Coli to do our work Why not just let living animals or plants make the proteins and isolate? Well, first of all there is the waste management problem! 40 kilos per day fed to an elephant produces a mess all its own! Some proteins cannot be isolated. For example, the adhesive proteins used by barnacles are secreted and soon crosslinked, becoming impossible to isolate. These “glue” proteins work in seconds, even under water. Some naturally-occurring research targets Elastin: protein of arteries and lungs that lasts a lifetime of stretching and contracting without degradation. Elastin is a molecular spring, and can undergo stretching of 300%. Viral spike proteins: fiber-like sequences at surfaces copied and can make liquid crystalline solutions and fibers with properties good enough for commercial use. Commercial interest Protein Polymer Technologies • Silk • Silk with fibronectin-like patches, useful to coat glassware used in cell culture. Available in >500g quantities. Allied-Signal • Collagen DuPont • Viral spike proteins Military interest U.S. Army: coiled coil (probably for fibers of good tensile and compressive strength) Making polymers that nature never intended to simulate structures nature makes all the time. Conventional Synthesis of the rigid, helical rod, PBLG=poly(benzylglutamate) O O n O O + NH2 N H -n CO2 O O H N H n O O CH2 NHC 6H13 Mw/Mn ~ 1.2 Bioengineering approach to PBLG • Code the nucleic acid for exact repeats of glutamic acid • Include an occasional aspartic acid to prevent enzymatic cleavage • Get E. Coli to make poly(glutamic acid)-co(aspartic acid) • Convert the acid groups to the benzyl ester Unnatural amino acids, including fluorinecontaining amino acids—a genetic engineering approach to low surface tension materials, hydrolytic stability, solvent resistance and low friction. One-piece protein-based nanodevices Functional part. The first one to be tried will mimic a phosphotriesterase enzyme that can detect and detoxify nerve toxins and some pesticides. Self-assembling base made of lysine or cysteine residues to attach to negative surfaces or gold, respectively. Part 2. Another aspect of bionano is measuring and emulating biomachines. The following material is taken from the website of the Stephen Block group at Stanford. This group is proficient in “grabbing” a large nanoparticle (e.g., latex) with light tweezers. That idea is associated with the name Ashkin and helped to win a Nobel Prize for Steven Chu (who initially did nothing related to polymers). Kinesin Cartoon of a kinesin experiment. The kinesin walks along the microtubule towards its plus-end, while being subjected to a retarding force by the optical trap. Cartoon of a kinesin experiment. The kinesin walks along the microtubule towards its plus-end, while being subjected to a retarding force by the optical trap. In order to track kinesin motion, we attach the molecules to microscopic beads. Kinesin itself is much too small to see in the optical microscope, so the beads serve as markers that can be tracked with very high precision (to 1 nm or better). The beads also act as "handles", through which we can apply force using an optical trap. Applying tension reduces Brownian motion of the bead, making it clear that kinesin moves in a stepwise fashion, in 8-nm increments (see below, and Svoboda et al., Nature 1993). For each 8-nm step, kinesin uses a single fuel molecule, hydrolyzing one ATP molecule into ADP and inorganic phosphate (Schnitzer and Block, Nature 1997). Kinesin Kinesin steps under constant forward load. The position of a kinesin motor in nanometers versus time at low ATP concentration. In this graph, the 8-nm steps that kinesin takes along the microtubule can readily be seen. This step size corresponds to the spacing of the tubulin dimers that make up the microtubule (Figure from Lang et al., Biophys. J. 2002). Energy Wells in which Kinesin Operates 3D cartoon of the effect of sideways loads. By using a 2D optical force clamp, we have been able to probe the multidimensional potential energy landscape that governs the kinetics of kinesin motion. DNA Helicase—Unwinds ds-DNA Static representation of a DNA helicase as it unwinds a piece of double stranded DNA. (Picture from Taekjip Ha, UIUC). Cartoon of a DNA unzipping experiment. The optical trap applies a force to the hairpin causing it to unzip. We are investigating the mechanical properties of nucleic acids by focusing in particular on hairpins. These structures consist of single strands of DNA or RNA whose ends are self-complementary, such that they loop back on themselves to form a duplex "stem" connected to a single-stranded loop (inset below). Hairpins not only provide a model system for studying DNA unzipping, but are also important in their own right (for example, they are a principal motif of secondary structure in RNA, and they play a role in gene transcription and regulation). The aim of our research is to understand, at a fundamental level, the factors that affect the stability of hairpins. We use a DNA "handle" to attach a hairpin to a small bead, which we can then manipulate with optical tweezers. As illustrated in the figure below, pulling on the bead with the tweezers applies a force to the hairpin, causing it eventually to unzip. By measuring the force at which the hairpin is pulled apart as well as the amount of time the hairpin spends in open and closed states at a given time, we are trying to map out the energetics of the hairpin while systematically varying parameters such as the neck length, neck sequence, and buffer conditions. Simultaneous fluorescence/tweezers The combination of single molecule fluorescence and optical tweezers in a hairpin unzipping experiment. The addition of the fluorescence signal allows us to better locate, both in space and time, a known structural event relative to an event observed with the optical tweezers (Figure from Lang et al. J. Biology 2003). RNA polymerase reading DNA Single molecule RNA Polymerase experiment using optical tweezers. RNAP is attached to a micron-sized polystyrene bead. The bead is then held in an optical trap equipped with an interference-based detection system. Tension is applied to the DNA, via an attachment to either the coverslip or a second bead (not shown). Transcription of RNAP affects the position of the bead. Jerky motion of RNA polymerase under tension Single E. coli RNA polymerase molecule transcribing under force. The position of the polymerase in base pairs (bp) is shown as a function of time. The motion of the polymerase is frequently interrupted by pauses of variable duration, between which the transcriptional velocity is constant. Watching a nanomotor at work As the RNAP transcribes along the DNA, the two beads get closer together. Here is a movie of a single RNAP transcribing along the DNA at 1 mM ATP,CTP,GTP,UTP sped up 30 times [RNAP Dumbell 30x.avi 178 KB].