* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Structural alignment wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Folding@home wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
List of types of proteins wikipedia , lookup
Alpha helix wikipedia , lookup
Protein domain wikipedia , lookup
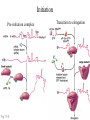
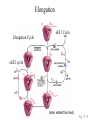
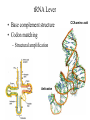
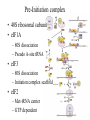
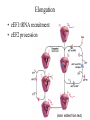
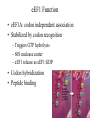
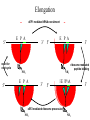
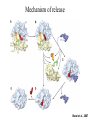
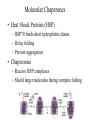
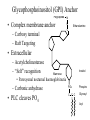
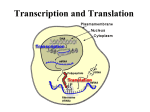
Protein Translation • Text Ch 3, 17 • Structure – Amino acids – Folding • Synthesis – – – – Pre-initiation Initiation Elongation Post-processing Protein structure Base • Amino acid H2N – CR – COOH – Amine – Side chain – Carboxylic acid • Amide backbone • Side chains Acid +H N 3 – CR – COO+H +H N 3 O- H – CR – CO – NH – CR – COO- – Polarity/charge – Size • Glycine “R” is –H • Tryptophan “R” is C9H8N with two rings 3-D structure • Solvent interaction – water – Hide the hydrophobes • Charge interaction – Acidic side chains (-) – Alkaline side chains (+) – Polar • Secondary structure – a-helix – b-sheet • Tertiary structure Protein Translation • • • • • Assembly of 5’-cap complex Annealing of ribosome t-RNA decoded polypeptide elongation Trafficking Co-translational modification – Sugars – Fatty acids – Chaperone mediated folding 80S Ribosome • Equivalent to RNA PolII or DNA Pold • Two major subunits: 40S & 60S APE 60S (large) subunit structrue 40S (large) subunit structrue tRNA docking 3 tRNA binding sites mRNA twisted through 40S Narrow peptide extrusion tunnel (spinnerette) Ban et al., 2000 Initiation Pre-initiation complex Fig 17-9 Transition to elongation Elongation eEF1 Cycle Elongation Cycle eEF2 cycle (note: edited from text) Fig 17-10 tRNA Lever • Base complement structure • Codon matching – Structural amplification Anticodon CCA-amino acid Pre-Initiation complex • 40S ribosomal subunit • eIF1A – 80S dissociation – Pseudo A-site tRNA • eIF3 – 80S dissociation – Initiation complex scaffold • eIF2 – Met-tRNA carrier – GTP dependent Initiation Complex • 43S Pre-Initiation Complex • mRNA – 7’methylguanosine (7mG) cap – eIF4 • eIF4G scaffold • eIF4E targeting • eIF4A ATP dependent helicase • Scanning – 5’ UTR structure 7mG cap eIF4E specifically binds 7mG cap Ribosome Assembly • 48S Initiation complex – Scans along mRNA for AUG – eIF5: eIF2 GAP – eIF5B: recruits 60S subunit • GTP hydrolysis displaces eIF5B • 60S subunit – Aminoacyl, peptidyl, exit docking sites – P site initially occupied by t-Met Elongation • eEF1:tRNA recruitment • eEF2 procession (note: edited from text) Elongation • eEF1a (bacterial EF-Tu) – GTP dependent – Recruits aa-tRNA to A site • P-protein bound to A-amino acid – Transitional tRNA state • eEF2 (bacterial EF-G) – GTP dependent – Displaces A-tRNA • Ribosomal Release Factor (rRF) eEF1 Function • eEF1A: codon independent association • Stabilized by codon recognition – Triggers GTP hydrolysis – 60S nuclease center – eEF1 release as eEF1:GDP • Codon hybridization • Peptide binding Translational accuracy • AA-tRNA synthesis • Codon matching – Structural amplification – 1 Å accuracy 2.5 Å H-bonds mRNA tRNA Anticodon CCA-amino acid Ribosome procession • eEF2 – Structurally similar to eEF1+tRNA – Displaces A/P site tRNA to P site – Prime A site • GTP hydrolysis – 60S nuclease center Elongation eEF1 mediated tRNA recruitment 5’ E P A 3’ 5’ E P A reset for next cycle ribosome mediated peptide binding NH3 5’ 3’ NH3 E P A 3’ 5’ EE PPAA eEF2 mediated ribosome procession NH3 NH3 3’ Elongation eEF2 40S eEF2 60S PDB IDs: 2XUX, 2XUY; 2XSY, 2XTG MMDB IDs: 111552, 111555 Ratje & al., 2010 Termination • eRF1 recruited to stop codon – – – – UAA, UAG, UGA Another structural analog of tRNA Breaks P-site peptide bond GTPase Mechanism of release Barat et al., 2007 Termination • eRF3 – eRF1 GAP – Dissociation of eRF1 by activating GTPase • eRF4 – 60S dissociation and recycling • Initiation factors – eIF3 Displaces P-site tRNA – eIF1 Post-translational Processing • Folding – Chaperone proteins – Endoplasmic reticulum • Trafficking – Subcellular localization – Targeting signals Protein folding • Energy minimization – Hydrophobic domains – Charge balance – Metallic complexes • Ribosome holds ~40 residues denatured • Spontaneous folding • Assisted folding Protein folding may be a stochastic search for the lowest energy configuration Molecular Chaperones • Heat Shock Proteins (HSP) – HSP70 binds short hydrophobic chains – Delay folding – Prevent aggregation • Chaperonins – Receive HSP complexes – Shield larger molecules during complex folding Subcellular trafficking • Posttranslational targeting to organelles • Cotranslational targeting to compartments – ER/Golgi – Signal sequence (Start/Stop) – Translocon Glycosylation • Co-translational addition of oligosaccharides – ER – Extracellular or membrane bound • • • • Negatively charged Highly hydrated Glycosaminoglycans (GAG) Binding/recognition – Synapse – ECM – Growth factor Acylation – fatty acid transfer • Myristic acid (C14:0) – NH3-Met-Gly– Co-translational amide bond with Gly • Palmitic acid (C18:0) – N-terminal, near TM domains – Thioester bond with Cysteine • Isoprenoids (C15:3/C20:4) – C-terminal CAAX box – Thioester bond with Cysteine – Cleavage of AAX • Membrane association • Acyl-chain coding of target membrane Saturated fatty acids Glycophosphoinositol (GPI) Anchor Polypeptide • Complex membrane anchor – Carboxy terminal – Raft Targeting N C Ethanolamine C • Extracellular – Acetylcholinesterase – “Self” recognition Inositol Mannose • Paroxysmal nocturnal haemoglobinuria – Carbonic anhydrase • PLC cleaves PO4 PO4 Phospho C C C Glycosyl Acyl