* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download No Slide Title
Fatty acid metabolism wikipedia , lookup
Genetic code wikipedia , lookup
Catalytic triad wikipedia , lookup
Point mutation wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Specialized pro-resolving mediators wikipedia , lookup
Butyric acid wikipedia , lookup
Biochemistry wikipedia , lookup
Citric acid cycle wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
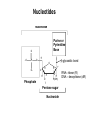
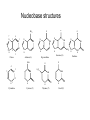
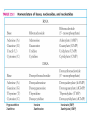
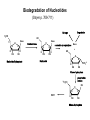
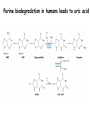
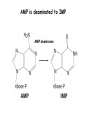
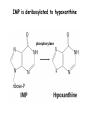
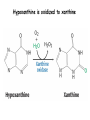
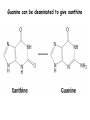
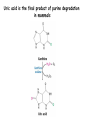
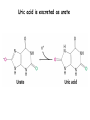
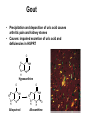
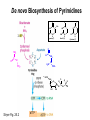
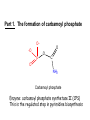
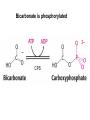
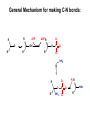
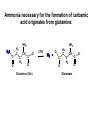
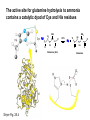
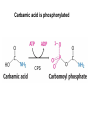
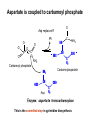
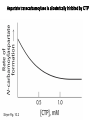
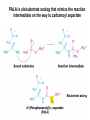
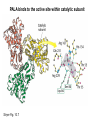
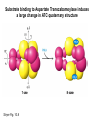
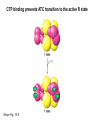
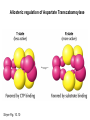
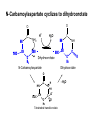
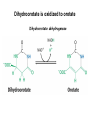
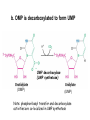
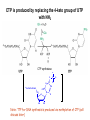
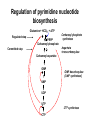
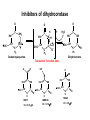
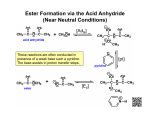
Biosynthesis of nucleotides Phar 6152 Spring 2004 Natalia Tretyakova, Ph.D. Required reading: Stryer’s Biochemistry 5th edition, p. 262-268, 693-712 (or Stryer’s Biochemistry 4th edition p. 238-244, 739-759) Tentative Lecture plan: Biosynthesis of Nucleotides 03-31 Introduction. Biological functions and sources of nucleotides. Nucleotide metabolism. 04-02 Biosynthesis of pyrimidine ribonucleotides. 04-05 Biosynthesis of purine ribonucleotides 04-07 Biosynthesis of deoxyribonucleotides. Inhibitors of nucleotide metabolism as drugs. 04-09 Review 04-12 Exam Biological functions and sources of nucleotides. Nucleotide metabolism Required reading: Stryer’s Biochemistry 5th Ed., p. 693-694, 709-711 Biological functions of nucleotides 1. 2. 3. 4. 5. a. b. c. Building blocks of nucleic acids (DNA and RNA). Involved in energy storage, muscle contraction, active transport, maintenance of ion gradients. Activated intermediates in biosynthesis (e.g. UDP-glucose, S-adenosylmethionine). Components of coenzymes (NAD+, NADP+, FAD, FMN, and CoA) Metabolic regulators: Second messengers (cAMP, cGMP) Phosphate donors in signal transduction (ATP) Regulation of some enzymes via adenylation and uridylylation Nucleotides Nucleotide Purine or Pyrimidine Base O 5' -O P b-glycosidic bond O O O- H 4' 3' H Phosphate H O 1' 2' H H(OH) Pentose sugar Nucleoside RNA- ribose (R) DNA – deoxyribose (dR) Nucleobase structures O O NH2 6 7 N 5 9N 4 N 1 N N N N O N NH NH NH 8 2 N H N H N H N N H N 3 Purine Adenine (A) Hypoxanthine NH2 O NH2 N Guanine (G) 3 O 6 H3C N N NH NH 2 N1 H Pyrimidine N H O Cytosine (C) N H Thymine (T) O N H Uracil (U) N H Xanthine 4 5 N H O O Hypoxanthine Xanthine Inosine Xanthosine Inosinate (IMP) Xanthylate (XMP) Two major routes for nucleotide biosynthesis dNTPs Stryer Fig. 25.1 dNTPs Nucleobase Products of Intracellular or dietary/intestinal degradation can be recycled via salvage pathways 1 and 2 (red) 1 2 Phosphoribosyl transferases involved in salvage pathway convert free bases to nucleotides adenine phosphoribosyl transferase Adenylate + PPi Adenine + PRPP hypoxanthine-guanine phosphoribosyl transferase Guanylate + PPi Guanine + PRPP Inosinate + PPi Hypoxanthine + PRPP 2- 2- O3PO CH2 O -O Base + O -O P O OH (HGPRT) O3PO CH2 O- Base O + PPi P O O OH 5-phosphoribosyl-1-pyrophosphate (PRPP) OH OH Biodegradation of Nucleotides (Stryer p. 709-711) Degradation Salvage 2- O3PO HO 5' Base O H H OH H OH H Nucleotide 5'-phosphate Nucleootidase O Base nucleoside phosphorylase H H OH H OH H Base HO O H H Nucleoside OPO32- H OH OH Ribose-1-phosphate phosphoribo mutase 2- O3PO OH O PRPP H H OH OH H Ribose-5-phosphate Nucleobase Products of Intracellular or dietary/intestinal degradation can be recycled via salvage pathways 1 and 2 (red) 1 2 Purine biodegradation in humans leads to uric acid AMP is deaminated to IMP AMP deaminase IMP is deribosylated to hypoxanthine phosphorylase Hypoxanthine is oxidized to xanthine Guanine can be deaminated to give xanthine Uric acid is the final product of purine degradation in mammals Uric acid is excreted as urate Deleterious consequences of defective purine metabolism • Gout (excess accumulation of uric acid) • Lesch-Nyhan syndrome (HGPRT null) • Immunodeficiency Gout • Precipitation and deposition of uric acid causes arthritic pain and kidney stones • Causes: impaired excretion of uric acid and deficiencies in HGPRT O N N H NH N Hypoxanthine O O NH N N H N Allopurinol NH N N H N H O Alloxanthine Lesch-Nyhan Syndrome • Caused by a severe deficiency in HGPRT activity • Symptoms are gouty arthritis due to uric acid accumulation and severe neurological malfunctions including mental retardation, aggressiveness, and self-mutilation • Sex-linked trait occurring mostly in males Lack of HGPRT activity in Lesch-Nyhan Syndrome causes a buildup of PRPP, which activates the synthesis of purine nucleotides hypoxanthine-guanine phosphoribosyl transferase Guanine + PRPP Hypoxanthine + PRPP Guanylate + PPi Inosinate + PPi •Excessive uric acid forms as a degradation product of purine nucleotides •Basis of neurological aberrations is unknown Immunodeficiency induced by Adenosine Deaminase defects AMP deaminase • Defects in AMP deaminase prevent biodegradation of AMP • AMP is converted into dATP • dATP inhibits the synthesis of deoxyribonucleotides by ribonucleotide reductase, causing problems with the immune system (death of lymphocytes, immunodeficiency disease) Summary: • Nucleotides have many important functions in a cell. • Two major sources of nucleotides are salvage pathway and de novo biosynthesis •Purine nucleotides are biodegraded by nucleotidases, nucleotide phosphorylases, deaminases, and xanthine oxidase. •Uric acid is the final product of purine biodegradation in mammals • Defective purine metabolism leads to clinical disease. Key concepts in Biosynthesis: Review •Committed step •Regulated step •Allosteric inhibitor •Feedback inhibition De novo Biosynthesis of Pyrimidines NH2 O N N O -O P NH O N O O -O O H O- P O OH OH O -O O H OH NH N O H H O H H OH O O OH OH P O H H OH H H H Required reading: Stryer’s Biochemistry 5th Ed., p. 262-267, 694-698 De novo Biosynthesis of Pyrimidines O NH2 O CH3 HN N HN O N H Thymine (T) O N H Cytosine (C) O N H Uracil (U) O NH2 C O -O C CH2 O H2N PO3- 2- O3PO CH COO- CH2 O -O O -O P O OH Stryer Fig. 25.2 dTTP OH O- P O O Part 1. The formation of carbamoyl phosphate O-O P O O C O NH2 Carbamoyl phosphate Enzyme: carbamoyl phosphate synthetase II (CPS) This is the regulated step in pyrimidine biosynthesis Bicarbonate is phosphorylated CPS : Phosphate is displaced by ammonia: CPS General strategy for making C-N bonds: C-OH is phosphorylated to generate a good leaving group (phosphate) General Mechanism for making C-N bonds: R R R' O- ADP R OH O R' ATP O R' P O ONH3 O R' Pi R O- R NH2 P O- NH2 O R' Ammonia necessary for the formation of carbamic acid originates from glutamine: NH2 H2 C H2N C O CH C H2 - C O O Glutamine (Gln) CPS H2 C - NH3 NH2 O C O CH C H2 Glutamate C O O- Structure of Carbamoyl phosphate synthetase II Stryer Fig. 25.3 The active site for glutamine hydrolysis to ammonia contains a catalytic dyad of Cys and His residues NH2 H2 C H2N C O CH C H2 C O- O Glutamine (Gln) Stryer Fig. 25.4 CPS NH3 NH2 H2 C - O C O CH C H2 Glutamate C O O- Carbamic acid is phosphorylated CPS Substrate channeling in CPS NH2 H2 C H2N C O CH C H2 - C O O Glutamine (Gln) Stryer Fig. 25.5 CPS NH3 NH2 H2 C - O C O CH C H2 Glutamate C O O- Carbamoyl phosphate supplies the C-2 and the N-3 of the pyrimidine ring O NH2 C O -O O C CH2 H2N PO3- dTTP CH COO- Part 2. The formation of orotate. Aspartate is coupled to carbamoyl phosphate O Asp replaces Pi Pi O-O P HN O O C O OOC NH2 Carbamoyl phosphate CH Asp CH NH2 COO C H2 Carbamoylaspartate NH2 -OOC C COOC H2 Enzyme: aspartate transcarbamoylase This is the committed step in pyrimidine biosynthesis Aspartate transcarbamoylase is allosterically inhibited by CTP Stryer Fig. 10.2 Allosteric regulation of Aspartate Transcarbamoylase Stryer Fig. 10.5 PALA is a bisubstrate analog that mimics the reaction intermediate on the way to carbamoyl aspartate Bisubstrate analog PALA binds to the active site within catalytic subunit Stryer Fig. 10.7 Substrate binding to Aspartate Transcabamoylase induces a large change in ATC quaternary structure Stryer Fig. 10.8 CTP binding prevents ATC transition to the active R state Stryer Fig. 10.9 Allosteric regulation of Aspartate Transcabamoylase Stryer Fig. 10.10 N-Carbamoylaspartate cyclizes to dihydroorotate O O HN OOC CH C + H H2O NH2 COO C Dihydroorotase H2 N-Carbamoylaspartate HN OOC C CH HN OOC CH N H H O C O- C H2 Tetrahedral transition state C C H2 Dihydroorotate O C NH - H2O O Dihydroorotate is oxidized to orotate Dihydroorotate dehydrogenase Part 3. The formation of UMP a. Orotate is phosphoribosylated to OMP O Pyrimidine phosphoribosyl transferase O - C HN OOC NH C O3PO CH2 O -O O -O P C C H - O O OH Orotate O- C COO OH OH Orotidylate (orotidine monophosphate, OMP) PRPP synthetase ATP O3PO OH N O OH AMP CH2 CH C O 5-phosphoribosyl-1-pyrophosphate (PRPP) 2- O CH2 P O PPi O3PO C HN O OH OH ribose-5-phosphate (from pentose phosphate pathway) b. OMP is decarboxylated to form UMP OMP decarboxylase (UMP synthetase) (OMP) (UMP) Note: phosphoribosyl transfer and decarboxylase activities are co-localized in UMP synthetase c.Phosphorylation of UMP gives rise to UDP and UTP: UMP kinase UMP + ATP UDP + ADP nucleotide diphosphate kinase UDP + ATP UTP + ADP CTP is produced by replacing the 4-keto group of UTP with NH2 CTP synthetase O O C4 N 4- O3PO3PO3PO-H2C O CH2 OH O- OCH C N O P C H OH Note: TTP for DNA synthesis is produced via methylation of CTP (will discuss later) Regulation of pyrimidine nucleotide biosynthesis Glutamine + HCO3- + ATP Regulated step PRPP Carbamoyl phosphate Committed step Carbamoyl aspartate OMP Carbamoyl phosphate synthetase Aspartate transcarbamoylase OMP decarboxylase (UMP synthetase) UMP UDP UTP CTP CTP synthetase Defects in de novo pyrimidine biosynthesis lead to clinical disease • Orotic acidurea – Symptoms: anemia, growth retardation, orotic acid excretion – Causes: a defect in phosphoribosyl transferase or orotidine decarboxylase O C HN OOC C - O3PO NH CH2 O -O -O P C C H Orotate O O O OH O- PPi P O O OH 5-phosphoribosyl-1-pyrophosphate (PRPP) – Treatment: patients are fed uridine U UMP UDP UTP UTP inhibits carbamoyl phosphate synthase II, preventing the biosynthesis and accumulation of orotic acid UTP inhibits carbamoyl phosphate synthase II, preventing the biosynthesis and accumulation of orotic acid Glutamine + HCO3- + ATP PRPP Carbamoyl phosphate Carbamoyl aspartate OMP UMP UDP UTP CTP Carbamoyl phosphate synthetase Drug inhibitors of pyrimidine biosynthesis Inhibitors of PRPP synthetase: 2- AMP ATP O3PO CH2 - O3PO O CH2 O -O O OH OH OH OH ribose-5-phosphate (from pentose phosphate pathway) NH2 OCH3 N N N N N N N N HO NH O H H HO NH O H H OH OH MRPP (MP) noncompetitive, Ki = 190 M -O P PRPP synthetase OH O H H H OH H OH ARPP (MP) noncompetitive, Ki = 430 M O- P O O Inhibitors of dihydroorotase O O C C HN OOC CH NH2 HN COO OOC C H2 O H+ H H2O N H CH C H2 HN NH OOC CH OO- C C Carbamoylaspartate C C H2 Dihydroorotate Tetrahedral Transition State -O O HS O SH P HC OOC CH O NH HC C N H HDDP Ki = 0.74 M O OOC CH HC NH OOC CH C N H O MMDHO Ki = 0.14 M CH C N H O MOAC Ki = 2.9 M Pyrimidine biosynthesis: take home message 1.Pyrimidines are synthesized by de novo and salvage pathways. 2. The pyrimidine ring is synthesized from pre-assembled ingredients (carbamoyl phosphate and aspartate) and then attached to the ribose. 3. Pyrimidine biosynthesis is tightly regulated via feedback inhibition (CTP synthetase, carbamoyl phosphate synthetase, aspartate transcarbamoylase) and transcriptional regulation (ATCase). 4. The mammalian enzymes are multifunctional (e.g. carbamoyl phosphate synthetase, UMP synthetase) and form multienzyme complexes to increase efficiency. 5. Drug inhibitors of pyrimidine biosynthesis are under development as potential antimicrobial and anticancer agents.