* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Proteolysis
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein domain wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Metalloprotein wikipedia , lookup
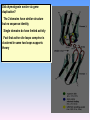
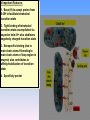
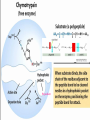
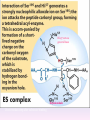
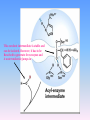
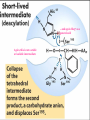
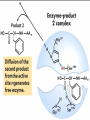
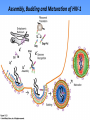
Proteolysis Proteases / Proteolysis Definitions: • – Proteases or peptidases are enzymes that catalyze the break down of peptide bonds in proteins/peptides • – Proteolysis is the process of controlled or uncontrolled degradation of proteins/peptides. Normal • • • • • • • Protein digestion Protein turnover Regulatory processes Apoptosis Cell differentiation Tissue remodeling Antigen presentation vs. Not Normal • • • • • • • • • • • Osteoporosis AIDS Alzheimer’s Angiogenesis Cancer Chagas’ disease Diabetes type 2 Diabetes & obesity Hypertension Peridontitis Epilepsy Serine Proteases • Enzyme and substrate become linked in a covalent bond at one or more points in the reaction pathway • The formation of the covalent bond provides chemistry that speeds the reaction • Serine proteases also employ general acid-base catalysis Serine Proteases Trypsin, chymotrypsin, elastase, thrombin (blood clotting cascade), subtilisin (bacterial protease), plasmin (degrades fibrin polymers of blood clots), TPA (cleaves plasminogen), Factor D • All involve a serine in catalysis - thus the name • Ser is part of a "catalytic triad" of Ser, His, Asp • Serine proteases are homologous, but sequence locations of the three crucial residues differ somewhat – Enzymologists agree, however, to number them always as His57, Asp102, Ser195 The Serine Proteases The amino acid sequences of chymotrypsinogen, trypsin, and elastase. • similar structures, yet different specificities Proteases: Members of the same family are usually evolutionarily related • Serine proteases • Aspartic proteases • Metalloproteases • Cysteine proteases Trypsin, Chymotrypsin, Elastase, Thrombin, Subtilisin, Factor D, Factor B, etc. Gastricsin, Cathepsin D, Renin, Pepsin, Chymosin, HIV protease, etc. Thermolysin,Carboxypeptidase, Matrix Metalloprotease (MMP) Papain, Calpain, Cruzipain, USP2a Catalytic Residues of Serine Proteases Ser: The only amino acid with a primary hydroxyl, often seen as a nucleophile in enzyme active sites. Can act as a hydrogen bond donor or acceptor. His: Histidine is the only amino acid with a pKa in the physiological range, hence often seen in active sites when donation or abstraction of a proton is needed. The protonated form shown has a pKa around 6 for the indicated proton , meaning it's mainly neutral at pH 7. Asp: Side chain pKa is about 4. One of two negatively charged amino acids. Did chymotrypsin evolve via gene duplication? The 2 domains have similar structure but no sequence identity Single domains do have limited activity Fact that active site loops comprise is clustered in same two loops supports theory • Chymotrypsin structure has two anti-parallel beta-barrel domains • Active site is formed by 2 loop regions from each domain. Serine Protease Mechanism A mixture of covalent and general acid-base catalysis • Asp102 functions only to orient His57 • His57 acts as a general acid and base • Ser195 forms a covalent bond with peptide to be cleaved • Covalent bond formation turns a trigonal carbon into a tetrahedral carbon • The tetrahedral oxyanion intermediate is stabilized by N-Hs of Gly193 and Ser195 4 Important Features: 1. Base (H) to accept proton from S-OH to facilitate tetrahedral transition state 2. Tight binding of tetrahedral transition state accomplished via oxyanion hole--H+ also stableizes negatively charged transition state 3. Nonspecific binding (due to main chain atoms H-bonding to main chain atoms of loop region in enzyme) also contributes to affinity/stabilization of transition state 4. Specificity pocket P1 pocket His57 acts as general base Oxyanion hole First reaction step after substrate binding The “intermediate” is highly unstable. It is a close mimic of the transition state. His57 now acts as a general acid Oxyanion hole Stabilizes the negative charge on the acyl “intermediate” This covalent intermediate is stable and can be isolated. However, it has to be Resolved to generate free enzyme and A water molecule jumps in His57 functions again as a general base Oxyanion Hole functions similar as in previous steps. …and again His57 as a general acid Again, this is not a stable or isolable intermediate Assembly, Budding and Maturation of HIV-1 • First drug developed for HIV-1 was AZT which inhibits Reverse Transcriptase (RT) • AZT is a nucleoside analog that on conversion to triphosphate in the cell, inhibits RT. • Unfortunately RT inhibitors only slow progression of HIV infection but do not stop it. 1. AZT-like drugs are toxic to bone marrow cells so can only taken as small doses. 2. RT unlikie other DNA polymerases cannot repair its mistakes so mutations eventually make the protein resistant to RT inhibitors. HIV -1 protease excises itself from gag-pol then cleaves other proteins on gag and gag-pol to produce its pathogenic mature form. If HIV-1 protease is inactivated either mutagenically or via inhibiotr, the virion remains noninfectious. The Aspartic Proteases • • • • Pepsin, chymosin, cathepsin D, renin and HIV-1 protease All involve two Asp residues at the active site These two Asp residues work together as general acid-base catalysts Most aspartic proteases have a tertiary structure consisting of two lobes (N-terminal and C-terminal) with approximate two-fold symmetry HIV-1 protease is a homodimer The Aspartic Proteases Most aspartic proteases exhibit a two-lobed structure. Each lobe contributes one catalytic aspartate to the active site. HIV-1 protease is a homodimeric enzyme, with each subunit contributing a catalytic Asp residue. Structures of (a) HIV-1 protease, a dimer, and (b) pepsin, a monomer. Pepsin’s Nterminal half is shown in red; the C-terminal half is shown in blue. Protease Inhibitors Block the Active Site of HIV-1 Protease HIV-1 protease complexed with the inhibitor Crixivan (red) made by Merck. The “flaps” that cover the active site are green; the catalytic active site Asp residues are violet. Protease Inhibitors Give Life to AIDS Patients • • • • Protease inhibitors as AIDS drugs If the HIV-1 protease can be selectively inhibited, then new HIV particles cannot form Several novel protease inhibitors are currently marketed as AIDS drugs Many such inhibitors work in a culture dish However, a successful drug must be able to kill the virus in a human subject without blocking other essential proteases in the body