* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chemical Biology I (DM)
Multi-state modeling of biomolecules wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Ligand binding assay wikipedia , lookup
Expression vector wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Paracrine signalling wikipedia , lookup
Biochemistry wikipedia , lookup
MTOR inhibitors wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Signal transduction wikipedia , lookup
Metalloprotein wikipedia , lookup
Drug discovery wikipedia , lookup
Interactome wikipedia , lookup
Protein purification wikipedia , lookup
Drug design wikipedia , lookup
Western blot wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Two-hybrid screening wikipedia , lookup
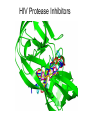
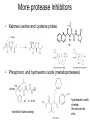
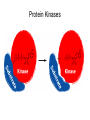
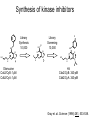
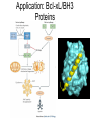
Chemical Biology 1 – Pharmacology 10-17-14 Methods for studying protein function – Loss of Function • 1. Gene knockouts • 2. Conditional knockouts pre-translational • 3. RNAi • 4. Pharmacology (use of small molecules to turn off protein function) Pharmacology • Advantages 1. Fast time scale 2. Only perturbs targeted sub-domains 3. graded dose response - tunability 4. Most drugs are small molecules • Disadvantage – Unlike genetic methods it is difficult to identify ligands that are highly selective for a target. Weiss WA, Taylor SS, Shokat KM. “Recognizing and exploiting differences between RNAi and small-molecule inhibitors.” Nat Chem Biol. 2007 Dec;3(12):739-44. Time Scale and Specificity Small molecules are subdomain specific Example: PAK1 Kinase Small molecules affect only one domain, while pre-translational methods remove the entire protein from the cell. Tunability Allows the amount of inhibition/activity that is necessary Reverse Chemical Genetics (Pharmacology) 1. Identify a protein target of interest – Develop an activity assay (enzymes) or a binding assay (protein-ligand interactions) to screen compounds 2. Test biased or unbiased panels of compounds against protein target of interest 3. Optimize your initial lead compound by making analogs (SAR) and by using any additional biochemical/structural information. In parallel, screen optimized analogs against other targets (selectivity) Major challenges • Druggability – Many proteins do not appear to make favorable interactions with drug-like small molecules Molecular Weight <900 Da Kd < 1 mM (∆G < -8.4 kcal/mol) No more than one or two fixed charges – Estimated that only ~10% of all proteins are druggable Hopkins and Groom, Nat Reviews Drug Disc, 2002 Major challenges • Selectivity – Finding selective agonists and antagonists is very challenging – Knowing which other proteins to counterscreen is difficult (easier for mechanism-based or enzyme family-directed ligands) In some cases, chemistry and genetics can be used to circumvent these problems. Knight ZA, Shokat KM. “Chemical genetics: where genetics and pharmacology meet. Cell. 2007 Feb 9;128(3):425-30.” Koh JT. “Engineering selectivity and discrimination into ligand-receptor interfaces.” Chem Biol. 2002 Jan;9(1):17-23. Review. Identification of small molecule inhibitors 2 classes – 1. Enzyme Inhibitors • Many effective strategies for identifying enzyme inhibitors. – 2. Protein-Protein Interaction Inhibitors • Difficult to identify potent inhibitors of proteinprotein interactions. Methods for discovering enzyme inhibitors • High throughput screening (parallel synthesis and combinatorial chemistry) • Mechanism-based (incorporate a functionality that is unique for an enzyme enzyme class (For example, proteases) • Privileged scaffolds (kinases, phosphodiesterases) • Transition state analogs Turk B.Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006 Sep;5(9):785-99. Aspartyl Protease Inhibitors HIV Protease Inhibitors INHIBITORS OF HIV-1 PROTEASE: A “Major Success of StructureAssisted Drug Design” Alexander Wlodawer, Jiri Vondrasek. Annual Review of Biophysics and Biomolecular Structure. Volume 27, Page 249-284, 1998 HIV Protease Inhibitors HIV Protease Inhibitors Resistance More protease inhibitors • Ketones (serine and cysteine proteases) • Phosphonic and hydroxamic acids (metalloproteases) transition state analog hydroxamic acids chelate the active site zinc Protein Kinases The human genome encodes 538 Protein kinases (483 are catalytically active) Kinase Inhibitors Almost all inhibitors that have been developed bind in the ATP pocket Synthesis of kinase inhibitors Library Synthesis 10,000 HN N N HO HN N N Olomucine Cdc2/CyB: 1µM Cdk2/CyA: 1µM N H R HN N N R' N H Cl Library Screening 10,000 N N N N R" HO N H N N Hit Cdc2/CyB: 340 pM Cdk2/CyA: 340 pM Gray et. al. Science (1998) 281, 533-538. Approved kinase inhibitors 28 small molecule kinase inhibitors are now in the clinic Gleevec (Imatinib) was the first clinically approved kinase inhibitor (2003) Protein-Protein Interaction (PPI) Inhibitors • Identification of potent PPI inhibitors is very challenging. In general, standard screening strategies don’t work. • Conversion of Peptides/Proteins to Small Molecules • Innovative new strategies are needed – for example, SAR by NMR Conversion of Peptides/Proteins to Small Molecules “SAR” by NMR Abbott Laboratories (Stephen Fesik) Fragment based approach - library of small compounds (several thousands) - build up larger ligands - n fragments may yield n2 compounds NMR: 15N-HSQC of target protein (2D NMR) Requirements 3D structure of target protein (NMR or other) large quantities of 15N-labeled protein (> 100 mg) NMR assignments of backbone N and HN atoms size of protein <40 kDa solubility: protein and ligands Principle start with known protein structure and 15N assignments 15N-HSQC of protein 15N-HSQC of protein plus ligand: identify shifted peaks map these on protein surface: binding site Shuker, S. B.; Hajduk, P. J.; Meadows, R. P.;Fesik, S. W. Science 1996, 274, 1531. “SAR” by NMR “SAR” by NMR 1. Screen 100-5000 low molecular weight (150 -300 MW) ligands to identify weak binders. HSQC perturbations identigy the site of binding 2. Screen for a second site of binding in the presence of the first ligand 3. Use structural information to design a linkage between the two identified ligands ∆G(linked ligand) = ∆G(fragment 1) + ∆G(fragment 2) + ∆G(linker) + ∆G(cooperativity) ∆G(linker) usually positive (entropic cost) ∆G(cooperativity) is a non-additive effect Application: Bcl-xL/BH3 Proteins First Site Ligands First Site Ligands Second Site Ligands Linked Inhibitor (Bcl-xL-ABT-737) NO2 H N S O O O NH S N N N Cl Nature 2005 Jun 2;Vol. 435(7042):p. 677-81. J. Med Chem. 2006 Jan 26;Vol. 49(2):p. 656-63. J Med Chem. 2006 Feb 9;Vol. 49(3):p. 1165-81