* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Overview
Protein moonlighting wikipedia , lookup
Epitranscriptome wikipedia , lookup
RNA interference wikipedia , lookup
List of types of proteins wikipedia , lookup
RNA silencing wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Genome evolution wikipedia , lookup
Non-coding RNA wikipedia , lookup
Gene desert wikipedia , lookup
Molecular evolution wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene expression wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene regulatory network wikipedia , lookup
Community fingerprinting wikipedia , lookup
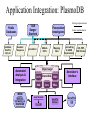
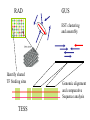
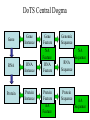
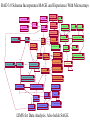
GUS Overview June 18, 2002 GUS-3.0 Genomics Unified Schema • Supports application and data integration • Uses an extensible architecture. • Is object-oriented even though it uses an underlying relational database management system (Oracle). • Warehouse instead of federation for local stable copy • Uses standards for bulk data exchange (e.g., MAGE) • Annotation GUS Usage – of genomes - gene models, sequence features – of genes - gene function, gene expression, gene regulation • Data mining – Develop algorithms and queryable resource • Publish – Map identifiers with other resources/ databases – URL for entry retrieval/ ad hoc queries in web interface GUS-3.0 Name Spaces GUS has 5 name spaces compartmentalizing different types of information. Namespace Domain Features Core Data Provenance Workflows Sres Shared resorurces Ontologies DoTS sequence and annotation Central dogma RAD Gene expresssion MIAME TESS Gene regulation Grammars Application Integration: PlasmoDB Public Databases GenBank, InterPro, GO, etc Existing implementation TIGR Sanger Stanford Genomic Sequence Automated Analysis & Integration Plasmodium Investigators GSSs & ESTs Annotation Object Layer DoTS Oracle/SQL TESS RAD WWW queries, browsing, & download Mapping Data Java Servlets & Perl CGI Core Future implementation microArray & SAGE Experiments Annotator’s Interface SRes GenePlot Software QTL,POP, SNP, Clinical GenePlot CD GUS Supports Multiple Projects AllGenes PlasmoDB EPConDB Java Servlets DoTS RAD TESS SRES Core Oracle RDBMS Object Layer for Data Loading Other sites, Other projects Main Aspects of GUS Development • Choice of development tools – Schema: • CREATE TABLE statements • Documentation plug-in: input is tab- delimited text • UML - Rational Rose, PowerDesigner – Code: CVS • Areas to emphasize – – – – – Plug-ins Work flow TESS Proteomics Images • Preferred type of user interface – JSP – PHP Data Integration Core Data Provenance • Ownership • Protection • AlgorithmsDoTS • Similarity • Versioning Genomic • Workflow Sequence SRes Ontologies • GO • Species • Tissue • Dev. Stage Transcribed Sequence Protein Sequence • Genes, gene models • STSs, repeats, etc • Cross-species analysis RAD Transcript • Characterize transcripts Expression • RH mapping • Library analysis • Cross-species analysis • DOTS • Domains • Function • Structure • Cross-species analysis •Arrays •SAGE •Conditions TESS Gene Regulation • Binding Sites • Patterns • Grammars Transcription factors up-regulated in acute myeloid leukemia with sequence similarity to c-fos and common promoter motifs RAD GUS EST clustering and assembly Identify shared TF binding sites TESS Genomic alignment and comparative Sequence analysis GUS Approach to Schema • Think objects – Parents and children – Subclassing with views • Views – Start with generic Imp table (e.g., NAFeatureImp) that contains base attributes plus generic attributes of various datatypes – Superclass view (e.g., NAFeature) just has base attributes – Subclass views (e.g., RNAFeature) have additional attributes using generic attributes • Strongly-typed – Tend to avoid “name-value” pairs DoTS Central Dogma Gene Instance Gene Feature RNA RNA Instance NA Feature RNA Feature Protein Protein Instance Gene Protein Feature AA Feature Genomic Sequence RNA Sequence Protein Sequence NA Sequence AA Sequence DoTS Schema Has Been Driven By Building Gene Indices Genomic Sequence Gene predictions GenScan/ HMMer, PHAT mRNA/EST Sequence SIM4 or BLAT Predicted Genes Merge Genes Clustering and Assembly DoTS consensus Sequences Gene/RNA cluster assignment Annotate DoTS Manual Annotation Tasks RNAs BLASTX Other computed annotation (EPCR, AssemblyAnatomyPercent, Index Key Words, SNP analysis) BLAST Similarities Gene Index framefinder translation BLASTP Functional predictions GO Functions Proteins PFAM, Smart, ProDom Protein Motifs DoTS Gene Indices Are Based on Clustering and Assembling ESTs Identify new sequences In GenBank and dbEST •Remove vector, polyA tails, ribosomal and poor quality sequences •Mask repeats with RepeatMasker “Quality” AssemblySequences •BLAST N vs self •BLASTN vs DoT S •Connected components analysis to form clusters Clusters of sequences (40 bp length, 92% identity) •Assemble clusters using CAP4 • update database Iterate to complete build -Extract consensus sequences GUS relational database -Block with RepeatMasker -BLAST N vs self -Cluster (95% identity, 75 bp overlap) -Assemble with CAP4 Annotation of DoTS consensus sequences -protein translations withframefinder -BLAST analyses vsnrdb, prodom and CDD -assign description and index keywords -GOFunction assignment -EPCR to generate radiation hybrid mapping -derive assembly -> anatomy mapping -alignment to genomic DNA -assignment to“Gene” clusters RAD 3.0 Schema Incorporates MAGE and Experience With Microarrays EXPERIMENTGROUP AnalysisOutput 0..* AnalysisImplementation LABEL 1 0..* 1 0..1 1 1 0..* AnalysisParameter GROUPFACTOR 0..* Analysis 1 0..* LABELEDEXTRACT 1 AnalysisInput BIOSAMPLE BIOSOURCE 0..* 1 BioMaterialImp ASSAYGROUPFACTOR 0..* 1 1 1 1 ARRAYANNOTATION 0..* ARRAY 0..* 1 0..* 0..* BIOMATERIALIMP 1 0..* RELATEDASSAY ASSAY 1 1 0..* 1 ASSAYLABELEDEXTRACT 1 0..* 0..* RELATEDACQUISITION ACQUISITION 0..1 BIOSOURCECHARACTERISTIC 0..* 0..* 1 0..* 0..* PROTOCOL 0..1 0..* 0..* CONTROL 1 1 1 0..* 1 ACQUISITIONPARAMETER 0..1 ONTOLOGYENTRY ELEMENTIMP 0..* TREATMENT 0..* 0..* ELEMENTANNOTATION 0..* 0..* CONTROLTYPE 0..* BIOMATERIALMEASUREMENT 1 0..* 1 1 0..* BioMaterialImp COMPOSITEELEMENTIMP 1 0..* 1 1 0..11 0..1 0..* 1 0..* RELATEDQUANTIFICATION QUANTIFICATION 0..* 0..1 1 0..* 1 COMPOSITEELEMENTANNOTATION QUANTIFICATIONPARAMETER 0..* 0..* 0..* ELEMENTRESULTIMP COMPOSITEELEMENTRESULTIMP 0..* PROCESSPARAMETER 0..1 0..* 1 0..* PROCESSTYPE PROCESS ProcessInput 1 1 0..* 0..1 0..* 0..* 0..* 0..* 1 PROCESSIMPPARAMETER 1 PROCESSIMPLEMENTATION ProcessOutput 1 0..* LIMS for Data Analysis. Also holds SAGE. 0..* Status of GUS Namespaces • Core – Tables exist, Workflow documented • Sres – Tables exist • DoTS – Tables exist, some documentation • RAD – Version 3.0 to include MAGE, experience • Pretty much complete – Tables exist, mostly documented • TESS – Tables ready but not created Schema Development • Releases on Sourceforge: – CREATE TABLE statements – Table dumps from Core::TableInfo, Core::DatabaseDocumentation – Gifs of ER diagrams • Adding tables between releases – In CVS tree? – Use message forum for discussion Documentation • Schema Browser looks at TableInfo • Plug-in – Populates DatabaseDocumentation – Input: Table\t\tDescription of table Table\tAttribute\tDescription of attribute GUS Schema Browser • http://www.cbil.upenn.edu/cgibin/GUS30/schemaBrowser.pl?db=GUS30 • Points at GUS30 on CBIL development database server (erebus). – Need to move? Maintain release view? • DoTS Tables: – – – – – Central dogma Evidence/ Similarity ProjectLink SequenceGroupImp/ SequenceGroupExperimentImp Plasmomap? • Other tables of interest?