* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Tertiary Structure
Catalytic triad wikipedia , lookup
Gene expression wikipedia , lookup
Magnesium transporter wikipedia , lookup
Interactome wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Biosynthesis wikipedia , lookup
Point mutation wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Western blot wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Structural alignment wikipedia , lookup
Biochemistry wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Proteolysis wikipedia , lookup
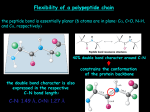
Tertiary Structure Chapter 3, 4, & 5 Tertiary Structure • Tertiary structure describes how the secondary structure units associate within a single polypeptide chain to give a three-dimensional structure. • Quaternary structure describes how two or more polypeptide chains associate to form a native protein structure (but some proteins consist of a single chain). • Tertiary structures can be divided into three main classes: a domain b domains a/b domains (Domain) Folds of Protein Common Features of Globular Proteins • The non-polar residues V, L, I, M and F largely occur in the interior of a protein, out of contact with the aqueous solvent. • The charged polar residues R, H, K, D and E are largely located on the surface of a protein in contact with the aqueous solvent. If they are located inside of a protein, they often have specific chemical function, such as promoting catalysis or participating in metal binding. • The uncharged polar groups S, T, N, Q, Y and W are usually on the surface but frequently occur in the interior of the protein, forming hydrogen bonds with other groups. • Globular protein cores are efficiently arranged with their side chains in relaxed conformations. The protein interior is efficiently packed (0.75). Alpha Domain: The Lone Helix • There are a number of examples of small proteins (or peptides), which consist of little more than a single helix. • A striking example is glucagon, a hormone involved in regulating sugar metabolism in mammals (as is insulin). Calcium-binding proteins (CaBPs) Binding affinities Kd (M) 10-3CaR Triggering Ca 10-4 Thermolysin 10-5 Calmodulin Ca2+-free Calmodulin (1cfc) Ca2+-binding Calmodulin (3cln) 10-7 Protease K Buffering Stabilizing 10-8 Calbindin D9k 10-9 α-Lactalbumin Parvalbumin Calbindin D9K (1b1g) Thermolysin (1tlx) Aequorin Calcium dependent protein from jellyfish Successfully expressed in the cell and can be targeted to specific cell subcompartments Requires a fluorescent cofactor, which is consumed, to be injected into the cell 1EJ3 Coil-coil a Helices • A coiled-coil a helix has a repetitive heptad amino acid sequence (transcription factor GCN4). Within each heptad the amino acids are labeled a-g. • The a helices in the coiled-coil are slightly distorted so that the helical repeat is 3.5 residues rather than 3.6, as in a regular helix. There is an integral repeat of seven residues along the helix. Packing of a Coil-coil • Every seventh residue in both a helices is a leucine, labeled “d”. • Due to the heptad repeat, the dresidues pack against each other along the coiled-coil. • Residues labeled “a” are also usually hydrophobic and participate in forming the hydrophobic core along the coiled-coil. Heptad Repeats. • Salt bridges can stabilize coiled-coil structures and are sometimes important for the formation of heterodimeric coiled-coil structures. The residues labeled “e” and “g” in the heptad sequence are close to the hydrophobic core and can form salt bridges between the two a helices of a coiled-coil structure, the e-residue in one helix with the g-residue in the second and vice versa. Helix-helix Packing • The side chains of an a helix are arranged in a helix row along the surface of the helix • They form ridges separated by shallow furrows or grooves on the surface. a helices pack with the ridges with one helix packing into the grooves of the other and vice visa. • Ridges can be formed by the sidechains separated by four residues with an angle of 25 º (i +4) and by 3 residues with an angle of 45 º (i +3) to the helical axis. Helix-helix Packing • By fitting the ridges of side chains from one helix into the grooves between side chains of the other helix and vice versa, a helices pack against each other. (a) Two a helices, I and II, with ridges from side chains separated by four residues marked in red and blue, respectively. In panel 4, the orientation of the helices has been rotated 50° in order to pack the ridges of one a helix into the grooves of the other. (b) In the red a helix, the ridges are formed by side chains separated by four residues and in the blue a helix by three residues. The a helices are rotated 20° in order to pack ridges into grooves, in a direction opposite that in (a). Knobs in Holes Model • The positions of the side chains along the surface of the cylindrical a helix is projected onto a plane parallel with the helical axis for both a helices of the coiled-coil. • The side-chain positions of the first helix, the "knobs," superimpose between the side-chain positions in the second helix, the "holes." Four-helix Bundles • Four-helix bundles frequently occur as domains in a proteins. • The arrangement of the a helices is such that adjacent helices in the amino acid sequence are also adjacent in the threedimensional structure. • Some side chains from all four helices are buried in the middle of the bundle, where they form a hydrophobic core. Four-helix Bundles • In cytochrome b562 (a) adjacent helices are antiparallel, whereas the human growth hormone (b) has two pairs of parallel a helices Dimeric RNA-binding Protein Rop RNA RNA • Each subunit of Rop comprises two a helices arranged in a coiled-coil structure with side chains packed into the hydrophobic core according to the "knobs in holes" model. • The two subunits are arranged in such a way that a bundle of four a helices is formed. • The RNA binding surface is located at the middle of the helices. The Globin Fold • This fold has been found in a large group of related proteins including myoglobin and hemoglobin. • The globin fold usually consists of eight alpha helices (AH). The two helices at the end of the chain are antiparallel, forming a helix-turn-helix motif, but the remainder of the fold does not include any characterized supersecondary structures. • These helices pack against each other with larger angles, around 50 ° between them than occurs between antiparallel helices (approximately 20°) so that the helices form a hydrophobic packet for the heme active site. The Globin Fold has been Preserved During Evolution • The 3D structures of globin proteins from different organisms (mammals, plants, and inserts) are solved and they share the same essential features of the globin fold. • The sequence homology is from 99 % to 16 %, which is very low. • How can amino acid sequences that are very different for proteins be very similar in their 3D structures? Evolution of Globins • Arthur Lesk & Cyrus Chothia in the UK have examined the residues that are structurally equivalent to positions in 9 known globin structures, that are involved in helix-heme contacts, and in the packing of the helices against each other. – There are a total of 59 positions preserved, 31 buried in the middle of protein and 28 in contact with the heme group. – There is no conserved sequences nor size-compensatory mutations in the hydrophobic core formed by the 31 a.a. – Conclusion: The evolutionary divergence of globins has been constrained primarily by an almost conservation of the hydrophobicity of the residues buried in the helix-helix and helix-heme contact. How do Proteins Adopt to Changes in the Size of Buried Residues? • The mode of packing for the a helices are the same in all the globin structures • The same types of packing ridges into grooves occur in corresponding a helices in all these structures. – The relative positions and orientations of the a helices change to accommodate changes in the volume of sidechains involved in the packing. – The structure of loop regions changes so that the movement of one helix is not transmitted to the rest of the structure to preserve the geometry of the heme pocket. Hemoglobin • The hemoglobin molecule is built up of four polypeptide chains: two a chains and two b chains. Each chain has a threedimensional structure similar to that of myoglobin: the globin fold. • In sickle-cell anemia, Glu 6 in the b chain of hemoglobin is mutated to Val, thereby creating a hydrophobic patch on the surface of the molecule. Sickle-cell Hemoglobin • Sickle-cell hemoglobin molecules polymerize due to the hydrophobic patch introduced by the mutation Glu 6 to Val in the b chain. Alpha/Beta Structure • Alpha/beta domains are found in many proteins. • The most regular and common domain structures consist of repeating beta-alpha-beta super-secondary units, such that the outer layer of the structure is composed of alpha helices packing against a central core of parallel beta sheets. These folds are called alpha/beta, or wound alpha beta. • There are three main classes of a/b proteins (b-a-b): – TIM barrel – Rossman Fold (open sheets) – Horseshoe fold TIM Barrel • A core of twisted parallel b strands arranged close together, like the staves of a barrel. • Here shows a closed barrel exemplified by schematic and topological diagrams for the enzyme triosephosphate isomerase. The b-a-b motifs are connected sequentially, and a helices are on the same side of the b sheet. Properties of TIM Barrel a/b-barrel has been found in more than 15 proteins. Most of them are enzymes with completely different amino acid sequences and different functions. Branched hydrophobic side chains dominate the core of this class of proteins. • In most a/b-barrel structures the eight b strands of the barrel enclose a tightly packed hydrophobic core formed entirely by side chains from the b strands. The core is arranged in three layers, with each layer containing four side chains from alternate b strands. The schematic diagram shows this packing arrangement in the a/b barrel of the enzyme glycolate oxidase. Methylmalonyl-coenzyme A Mutase • One exception, the inside of the barrel methylmalonylcoenzyme A mutase is lined by small hydrophilic side chains (serine and threonine) from the b strands, which creates a hole in the middle where one of the substrate molecules, coenzyme A (green), binds along the axis of the barrel from one end to the other. Coenzyme A (CoA) a/b Barrel Active Site • The active site in all a/b barrels is a pocket formed by the loop regions that connect the carboxy ends of the b strands with the adjacent a helices (a). • A view from the top of the barrel of the active site of the enzyme RuBisCo (ribulose bisphosphate carboxylase), which is involved in CO2 fixation in plants (b). Open a/b Sheet • Open twisted b sheet is surrounded by ahelices on both sides of the b-sheet. • Examples of different types of open twisted a/b structures (a) the FMN-binding redox protein Flavodoxin and (b) the enzyme adenylate kinase, which catalyzes the reaction AMP + ATP <=> 2ADP. Active Site of Open a/b Sheet • There are always two adjacent bstrands on opposite sides of a bsheet. One of the loops from one of these two b-strands goes above the b-sheet, whereas the other loop goes below, which creates a crevice outside the edge of the b-sheet between two loops. • Almost all binding sites in this class of proteins are located in crevices at the carboxy end of the b sheet. •A schematic view of the active site of tyrosyl-tRNA synthetase. Tyrosyl adenylate, the product of the first reaction catalyzed by the enzyme, is bound to two loop regions: residues 38 - 47, which form the loop after b strand 2, and residues 190 - 193, which form the loop after b strand 5. The tyrosine and adenylate moieties are bound on opposite sides of the b sheet outside the carboxy ends of b strands 2 and 5. a/b–horseshoe Fold a/b –horseshoe Fold is formed by amino acid sequences that contain repetitive regions of a specific pattern of a helices and b-strands. The b strands form a curved parallel b sheet with all the a helices on the outside. Schematic diagram of the structure of the ribonuclease inhibitor. The molecule, which is built up by repetitive b-loop-a motifs, resembles a horseshoe with a 17-stranded parallel b sheet on the inside and 16 a helices on the outside. Leucine-rich Motifs • Consensus amino acid sequence and secondary structure of the leucine-rich motifs of type A and type B. “X” denotes any amino acid; “a” denotes an aliphatic amino acid. Conserved residues are shown in bold in type B. • In the ribonuclease inhibitor, leucine residues 2, 5, and 7 from the b strand pack against leucine residues 17, 20, and 24 from the a helix as well as leucine residue 12 from the loop to form a hydrophobic core between the b strand and the a helix. Beta Structures • Anti-parallel b strands are usually arranged in two b-sheets that pack against each other and form a distorted barrel structure, the core of the structure. • Depending on the way the bstrands around the barrel are connected along the polypeptide chain, they can be divided into four major groups: Up-and-down barrel superoxide dismutase (SOD) Greek Key barrel Jelly roll barrel b-helix Up-and-down Barrel • Schematic and topological diagrams of an up-and-down b barrel. • The eight b strands are all antiparallel to each other and are connected by hairpin loops. • Beta strands that are adjacent in the amino acid sequence are also adjacent in the threedimensional structure of up-and-down barrels. Retinol-binding Protein (RBP) • The structure of human plasma retinol-binding protein (RBP) is an up-and-down b barrel. A retinol molecule, vitamin A (yellow), is bound inside the barrel, between the two b sheets, such that its only hydrophilic part (an OH tail) is at the surface of the molecule. Green Fluorescent Protein from Aequorea victoria (jellyfish) 11-stranded b-barrel with central ahelix chromophore formed during folding (from residues S65, Y66, G67) is contained in the middle of the barrel contains 1 Trp, 10 Tyr Excitation 395 nm and 475 nm Emission 506 nm http://www.plantsci.cam.ac.uk/Haseloff/GFP/GFPbackgrnd.html Amino Acid Sequence Reflects b Structure • Amino acid sequence of b strands 2, 3, and 4 in human plasma retinol-binding protein. • The sequences are listed in such a way that residues which point into the barrel are aligned. • These hydrophobic residues are shown by arrows and are colored green. The remaining residues are exposed to the solvent. Greek Key Motifs • This motif is formed when one of the connections of four antiparallel b strands is not a hairpin connection. • The motif occurs when strand number n is connected to strand n + 3 (a) or n - 3 (b) instead of n + 1 or n - 1 in an eight-stranded antiparallel b sheet or barrel. The two different possible connections give two different hands of the Greek key motif. • In all protein structures known so far, only the hand shown in (a) has been observed. The Fold of IgG Domains • Beta strands labeled A-G of the constant and variable domains of immunoglobulins have the same topology and similar structures. There are two extra b strands, C' and C'' (red) in the variable domain. The loop between these strands contains the hyper-variable region CDR2. The remaining CDR regions are at the same end of the barrel in the loops connecting b strands B and C and strands F and G. Gamma Crystallin Domain • The domain structure of g-crystallin is built up from two b sheets of four antiparallel b strands, sheet 1 from b strands 1, 2, 4, and 7 and sheet 2 from strands 3, 5, 6, and 8. • It is obvious that the b strands are arranged in two Greek key motifs, one (red) formed by strands 1 - 4 and the other (green) by strands 5 - 8. Complete g-crystallin Molecule • The two domains of the complete molecule have the same topology; each is composed of two Greek key motifs that are joined by a short loop region. • There is a greater amino acid sequence homology between the domains than the motifs within each domain, suggesting that the four Greek Key motifs in gcrystallin are evolutionarily related by gene duplication and fusion. Jelly Roll Motifs • The eight b strands are drawn as arrows along two edges of a strip of paper. The strands are arranged such that strand 1 is opposite strand 8, etc.. • The b strands follow the surface of the barrel and the loop regions provide the connections at both ends of the barrel. Two-sheet b helix. • The two parallel b sheets are colored green and red, the loop regions that connect the b strands are yellow. • Each structural unit is composed of 18 residues with 9 consensus sequence Gly-Gly-X-Gly-X-AspX-U-X forming a b-loop-b-loop structure, where U is a large hydrophobic residue, often Leu. • Each loop region contains six residues of sequence Gly-Gly-XGly-X-Asp where X is any residue. Calcium ions are bound to both loop regions. Extracellular bacterial proteinase Three-sheet b Helix • As shown in (a), two of the b sheets (blue and yellow) are parallel to each other and are perpendicular to the third (green). In (b), each structural unit is composed of three b strands connected by three loop regions (labeled a, b and c). • Loop a (red) is invariably composed of only two residues, whereas the other two loop regions vary in length. Two pathways whereby enveloped viruses enter cells. • Schematic comparison of the VV 14-kDa protein, HIV gp41, Mo-MLV TM, and influenza HA2 structures. The four proteins form threestranded coiled-coil structures involving a central -helix. For all of them, the hydrophobic fusion peptide would be immediately amino terminal to the oligomerization domain, although for the VV 14-kDa protein the peptide implicated in this process has not been defined yet (22). For the 14-kDa protein an anchoring domain is indicated instead of a transmembrane region of the C terminus. Except for the influenza HA2, these fusion proteins have cysteine residues at the end of the coiled-coil region. Virus Fusion Proteins • Common structural elements between MoMLV TM, HIV gp41 and influenza HA2. The top panel shows schematic maps of the MoMLV, HIV and influenza sequences. For each, the position of the coiled coil is shown in red, and the position of the supporting structures is shown in blue. The bottom panel shows the structures of MoMLV TM residues 45–98 [Fass et al 1996], HIV gp41 residues 546– 581 [Chan et al 1997], and influenza HA2 residues 40–129 [Bullough et al 1994]. The interior coiled coils are shown in red, and the exterior supporting structures are shown in blue. The Globular Head of the Hemagglutinin Subunit is a Distorted Jelly Roll Structure b strand 1 contains a long insertion, and b strand 8 contains a bulge in the corresponding position. Each of these two strands is therefore subdivided into shorter b strands. The loop region between b strands 3 and 4 contains a short a helix, which forms one side of the receptor binding site (yellow circle). Structure of the HIV-1 gp41 core Suntoke, T. R. et al. J. Biol. Chem. 2005;280:19852-19857 • • A model of the fusion core structure of SARS-CoV spike protein: (a) sequence alignment of HR1 and HR2 region between MHV and SARS-CoV spike proteins; (b) model of fusion core structure of SARSCoV S protein. A surface map showing the hydrophobic grooves on the surface of three central HR1 is presented on the right side. Three HR2 helices pack against the hydrophobic groove in an antiparallel manner (left side). The helical regions in HR2 extended regions could be observed clearly. The figure on the right side shows the deep and relatively shallow grooves on the surface of the central HR1 coiled coil. The helical region of HR2 just packs against the deep groove and the extended region packs against shallow grooves. The deep groove would be an important target site for the fusion inhibitor design. Fig. 1. (A) Schematic representation of the coronavirus spike protein structure Bosch, Berend Jan et al. (2004) Proc. Natl. Acad. Sci. USA 101, 8455-8460 Copyright ©2004 by the National Academy of Sciences