* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Peptide bonds, polypeptides and proteins printable pdf
Chloroplast DNA wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Magnesium transporter wikipedia , lookup
Interactome wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Point mutation wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Metalloprotein wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Peptide synthesis wikipedia , lookup
Biosynthesis wikipedia , lookup
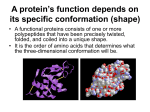
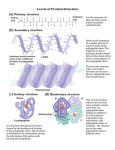
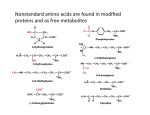
Biofundamentals - Peptide bonds and polypeptides 9/27/08 10:44 AM Peptide bonds, polypeptides and proteins printable pdf We have already mentioned proteins, since there are few biological processes that do not rely on them. Proteins derive their name from the ancient Greek sea-god Proteus who, like your typical sea-god, could change shape. The name acknowledges the many different properties and functions of proteins. Proteins can act as catalysts and regulators of chemical reactions – we have already seen how proteins act to regulate transport across membranes. Proteins control the expression of genes, how genes respond to internal and external signals, and the replication of the genetic material. They can act as structural components, determining both the shape and mechanical properties of cells and tissues. They can be motors, responsible for movements within cells and the movement of cells, tissues and the organism as a whole. Proteins are composed of α-amino acids linked together by peptide bonds into polypeptide chains. An amino acid is characterized by an amino group (NH 2 ) and a carboxylic acid group (-COOH) linked to a carbon, known as the α-carbon. Also attached to the αcarbon is a H and various R-groups or "sidechains". The four groups attached to the α-carbon are arranged at the vertices of a tetrahedron. If all four groups attached to the α-carbon are different from one another, as they are in all amino acids except glycine, the amino acid can exist in two possible stereoisomers, which are known as enantiomers. Enantiomers are mirror images of one another and are termed the L- and D- forms. Only L-type amino acids are found in proteins, even though there is no obvious reason that proteins could not have also been made using both types of amino acids, or using D-amino acids. It is not that D-amino acids do not occur in nature, or in organisms, they do. They are found in biomolecules, such as the antibiotic gramicidin, which is composed of file:///Users/klymkowsky/Documents/WebSites/virtual/Biofundamentals/lectureNotes/Topic3-4_Peptide%20bonds.htm Page 1 of 8 Biofundamentals - Peptide bonds and polypeptides 9/27/08 10:44 AM are found in biomolecules, such as the antibiotic gramicidin, which is composed of alternating L-and D-type amino acids – however gramicidin is synthesized by a different process than that used to synthesize proteins. It appears that the universal use of L-type amino acids is yet another example of the evolutionary relatedness of organisms (it appears to be a homologous trait). Even though there are hundreds of different amino acids known, only 19 amino acids and one imino acid, proline, are found in proteins. These amino acids differ in their R-groups. Some of these R-groups are highly hydrophobic, some are hydrophilic, some are positively or negatively charged. The different R-groups provide proteins with a range of chemical properties. In theory, what limits are there to the structure of an R-group? Is the use of a common set of amino acids among living organisms an analogous or a homologous trait? Building a polypeptide: Polymers are chains of subunits, monomers, linked together by chemical bonds. The bond between two amino acids is known as a peptide bond. Two amino acids, joined together by a peptide bond, is known as a dipeptide. An amino acid polymer is known as a polypeptide. A polypeptide can be composed of 20 different possible monomers that can, in theory, be linked together in any imaginable order. The formation of a peptide bond involves what is known as a condensation reaction. file:///Users/klymkowsky/Documents/WebSites/virtual/Biofundamentals/lectureNotes/Topic3-4_Peptide%20bonds.htm Page 2 of 8 Biofundamentals - Peptide bonds and polypeptides 9/27/08 10:44 AM In a condensation reaction two molecules are joined together and a molecule of water is released. In a hydrolysis reaction, the reverse occurs. The addition of a water molecule is associated with splitting the original molecule into two. In the case of both nucleic acids and polypeptides, polymer assembly involves a condensation reaction (with the release of water), while polymer disassembly into monomers involves a hydrolysis (with water as a reactant). Polypeptides can be gives rise to very large numbers of possible polypeptides; there are 20100 different polypeptides that are 100 amino acid residues long. During the synthesis of a polypeptide, an amino acid is attached to the C- or carboxyl terminus of the existing chain. This generates an unbranched, linear polymer. The peptide bond has a number of characteristics that are critical in determining how polypeptides behave. Although drawn as a single bond, the peptide bond behaves more like a double bond. While there is free rotation around a single bond, rotation around a peptide bond is constrained. Moreover, the carbonyl oxygen (C=O) acts as a H-bond acceptor, while the amino hydrogen (-N-H) acts as a H-bond donor. file:///Users/klymkowsky/Documents/WebSites/virtual/Biofundamentals/lectureNotes/Topic3-4_Peptide%20bonds.htm Page 3 of 8 Biofundamentals - Peptide bonds and polypeptides 9/27/08 10:44 AM Proline is not an amino acid, but rather an imino acid. Proline peptide bonds are found in the cis configuration ~100 times as often as those between other amino acids. There is no H-bond donor in the proline peptide bond and the presence of proline leads to a bend or kink in the polypeptide chain. Consider a condensation reaction; how would the reaction be effected if we removed water from the system? Are there any theoretical constraints on the order of monomers in a polypeptide? Why does it matter that rotation around a peptide bond is "constrained"? Water, polypeptide synthesis and folding: Were it not for the presence of hydrophobic R-groups, all polypeptides would assume an extended configuration in water. H-bond donors and acceptor groups in polypeptide backbone would form Hbonds with each other and with water molecules. More typical polypeptides have all of the different R-groups in various proportions. This sequence of amino acid residues, that is what is left of the amino acid once it is part of the polypeptide, is known as the primary structure of the polypeptide. We write the amino acid sequence of a polypeptide from its N- or amino terminus to its C- or carboxyl terminus, with the N-terminus to the left and the C-terminus to the right. A number of the amino acid R-groups are hydrophobic. Their presence makes an extended configuration for the polypeptide energetically unfavorable. Very much like the process by which lipids self-assemble to form micelles and bilayers, a typical polypeptide in aqueous solution will collapse onto itself in order to file:///Users/klymkowsky/Documents/WebSites/virtual/Biofundamentals/lectureNotes/Topic3-4_Peptide%20bonds.htm Page 4 of 8 Biofundamentals - Peptide bonds and polypeptides 9/27/08 10:44 AM bilayers, a typical polypeptide in aqueous solution will collapse onto itself in order to remove as many of these hydrophobic residues from contact with water as possible. It is a basic assumption of structural biology that the final folded state of a polypeptide is determined by the sequence of amino acids along its length, and that this final structure is the state of lowest, or close to the lowest, energy. Protein structure is commonly presented in a hierarchical manner. While this is an over-simplification, it is a good place to start. As it is being synthesized, the process known as translation, the polypeptide chain begins to fold. We can think of the folding process as a walk across an energy landscape. The goal is to find the lowest point in the landscape, the energy minimum of the system. This is generally assumed to be the native or functional state of the polypeptide. The first step is the movement of hydrophobic R-groups out of contact with water. This drives the collapse of the polypeptide into a compact and dynamic "molten globule". The path to the native state is not necessarily a smooth or predetermined one. The folding polypeptide can get "stuck" in a local energy minimum; there may not be enough energy for it to get out again. If a polypeptide gets stuck, there are mechanisms to unfold it and let it try again to reach its native state.This process of partial unfolding is carried out by proteins known as chaperones. What happens to a typical protein if you place it in a hydrophobic solvent? file:///Users/klymkowsky/Documents/WebSites/virtual/Biofundamentals/lectureNotes/Topic3-4_Peptide%20bonds.htm Page 5 of 8 Biofundamentals - Peptide bonds and polypeptides 9/27/08 10:44 AM What would be the structure of a polypeptide if all of its R-groups were hydrophilic? What is the basic characteristic of the molten globule configuration of a newly folded polypeptide? How might a chaperone recognize a misfolded polypeptide? Some amino acid R-groups contain carboxylic acid or amino groups, and so also act as weak acids and bases. Depending on the pH of the solution they are in, these groups may be protonated or unprotonated. Whether a group is charged or uncharged can have a dramatic effect on the structure, and therefore the activity, of a protein. By regulating pH, an organism can modulate the activity of specific proteins. There are, in fact, compartments within eukaryotic cells that are maintained at low pH in part to regulate protein activity. Why can changing the pH of the solution a protein finds itself in alter the protein's structure. Secondary structure features: All polypeptides share a common backbone structure made up of peptide bonds. It is therefore not surprising that there are common patterns in polypeptide folding. The first of these, the α- heIix, was discovered by Linus Pauling and Robert Corey, and reported in 1951. . This was followed shortly thereafter by the structure of the β-sheet. The forces that drive the formation of the α-helix and the β-sheet will be familiar. They are they same forces that underlie water structure. Linus at age 5 file:///Users/klymkowsky/Documents/WebSites/virtual/Biofundamentals/lectureNotes/Topic3-4_Peptide%20bonds.htm Page 6 of 8 Biofundamentals - Peptide bonds and polypeptides 9/27/08 10:44 AM In an α-helix and a β-sheet, all of the possible H-bonds involving the peptide bond's donor and acceptor groups are formed. This stabilizes the structure. In an α-helix , the R-groups point outward from the long axis of the helix. In β-sheets, the Hbonds between the peptide bond donor and acceptor groups are made between either parallel or antiparallel polypeptide chains. These are anti-parallel ßsheets The R-groups point out of and into the plane of the sheet. The key factor that determines the specific details of polypeptide folding is the interaction between R-groups, not only with one another but with water and dissolved ions. In a water soluble polypeptide, most but not necessarily all of the surface R-groups will be hydrophilic. Hydrophobic groups will tend to be buried in the polypeptide's interior. Some polypeptides are inserted into membranes. In these polypeptides, hydrophobic R-groups on the surface of the folded polypeptide will interact with the hydrophobic interior of the lipid bilayer. Other factors in protein structure: In addition to hydrophobic/hydrophilic effects, ionic interactions between acidic and basic R-groups, van der Waals interactions and H-bonds play an important role in determining the tertiary or three dimensional structure of the polypeptide. Proteins can include non-amino acid-based components, known generically as cofactors. A protein minus its cofactors is known as the apoprotein. Together with its cofactors, it is known as the holoprotein. Generally, without its cofactors, a protein is inactive and often unstable. file:///Users/klymkowsky/Documents/WebSites/virtual/Biofundamentals/lectureNotes/Topic3-4_Peptide%20bonds.htm Page 7 of 8 Biofundamentals - Peptide bonds and polypeptides 9/27/08 10:44 AM Cofactors can range in complexity from a single metal atom to quite complex molecules, such as vitamin B12 . For example, the retinal group of bacteriorhodopsin is a co-factor. A protein is a functional entity. In many cases a protein is composed of multiple polypeptides. A protein that contains multiple polypeptides, whether of the same or different types, is said to have a quaternary structure. A protein with two polypeptide subunits. There are also higher levels of polypeptide organization. For example, a specific polypeptide may be a component of a number of different proteins. Summarize the differences in structure between a protein that is soluble in the cytoplasm and one that is buried in the membrane. Why might proteins that require co-factors misfold in the absence of the co-factor? Any ideas about why cofactors might be necessary? How could surface hydrophobic amino acid R-groups facilitate protein-protein interactions. Use Wikipedia | revised 27-Sep-2008 file:///Users/klymkowsky/Documents/WebSites/virtual/Biofundamentals/lectureNotes/Topic3-4_Peptide%20bonds.htm Page 8 of 8