* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download NK Cells
Immune system wikipedia , lookup
Molecular mimicry wikipedia , lookup
Lymphopoiesis wikipedia , lookup
Adaptive immune system wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Psychoneuroimmunology wikipedia , lookup
Immunosuppressive drug wikipedia , lookup
Cancer immunotherapy wikipedia , lookup
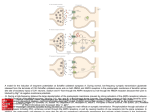
NK Cells Experimental Basis of Immunology January 17, 2007 W.H. Chambers, Ph.D. G.17e Hillman Cancer Center 623-3218 [email protected] I. Introduction • Natural Killer (NK) Cells were first described in the early 1970’s by R. Herberman; R. Kiessling; and G. and E. Klein • Defined as a functional entity, i.e. cell capable of recognizing and killing tumor cells without prior exposure • Represent a component of the non-adaptive immune system • Defined in the early 1980’s as having a large granular lymphocyte (LGL) morphology (Reynolds, et al., 1981) • Represent a heterogeneous population of cells with diverse functions • Can be best defined phenotypically as CD3-, CD16+, CD56+, CD122+, CD158+, CD161+ Innate capacity of lysis Large granular lymphocytes CD3-, CD16+, CD56+, CD122+, CD158+, CD161+ II. Pathway of NK Cell Differentiation: Topics • Differentiation of NK cells in the fetus • Differentiation of NK cells in adults • Terminal differentiation of mature NK cells NK Cell Differentiation • Derive from, and require normal, intact bone marrow for functional maturation • Represent one of the major lymphocyte populations [T, B, NK, NK-T] – ~5% of cells among PBLs • Present in athymic [nude] mice and rats • Present in scid mice, and in RAG-1 and RAG-2 knockout mice • Can be distinguished from other lymphocytes by the absence of clonally distributed, receptors derived via gene rearrangements NK Progenitors: Fetus Fetal thymus p-T Fetal liver HSC Fetal blood CLP c-T/NKP c-kit+ Thy-1CD25CD161c- c-kit+ Thy-1CD25CD161cCD19B220lo Modified from Lian and Kumar, 2002 p-T/NK c-kit+ Thy-1+ CD25CD161c+ p-T/NK c-kit+ Thy-1+ CD25CD161c+ T c-kit+ Thy-1+ CD25+ CD161c- p-NK c-kitThy-1+/CD25CD161c+ . . .. NK Differentiation of NK Cells: In Vitro Requirements for Growth and Maturation - Stroma from normal animals estrogen- or strontium-treated mice have functionally impaired NK cells stroma from LTa-/- mice have functionally impaired NK cells - Cytokines for growth and Differentiation c-kit ligand; IL7; Flt3 ligand [stem cell factor]; IL15 - Cytokines and direct contact with stroma are required for differentiation of phenotypically and functionally mature NK cells – LY49- NK cells develop in cultures with cytokines but no stroma NK Progenitors: Adult Bone marrow stroma HSC CLP NKP p-NK Linc-kitlo Thy-1IL7R+ Sca-1lo CD122+ CD161cCD49b- CD122+ CD161c+ CD49b- . . .. .. NK NK .. CD122+ CD161c+ CD49b+ Ly49+ CD122+ CD161c+ CD49b+ Ly49+ Thymus . . .. p-NK NK p-T T p-T/NK Modified from Lian and Kumar, 2002 T HSC MY X pB CLP Bone Marrow T/NKP NKP pT . . .. NK . . .. Periphery NK NK . .. . CTX . . .. NK Enhanced CTX NK Stimulus NK Cytokines: IFNg GM-CSF TNFa Modified from Yokoyama, et al, 2004 NK NK NK Progenitors: Adult Periphery Cytokines: IFNg GM-CSF TNFa . . .. NK Stimulus: Hrs IL2 IL12 IL15 IL23 IL27 IFNa, -b NK NK Enhanced CTX NK NK NK Progenitors: Adult Periphery . . .. NK Stimulus: Days IL2 IL12 IL15 IL23 IL27 IFNa, -b NK NK Cytokines: IFNg GM-CSF TNFa NK NK Enhanced CTX, with broader specificity Proliferation Knockouts/Transgenics: Transcription Factors Gene Deleted Ikaros Effect NK cells absent PU.1 NK cell number decreased, normal lytic function NK cell number decreased, Barton, 1998 decreased lytic function NK cells decreased or absent, Yokota, 1999 reduced lytic function Ikawa, 2001 Altered acquisition of Ly49sHeld, 1999 Kunz,2001 NK cell number decreased, Duncan, 1996 lytic function impaired Ogasawara, 1998 NK cell number decreased, Lohoff, 2000 lytic function impaired Ets-1 Id2 TCF-1 IRF-1 IRF-2 Reference Georgopoulos, 1994 Wang, 1996 Colucci, 2001 Knockouts/Transgenics: Receptors Gene Deleted LTbr LTa1b2 Effect NK cells severely decreased NK cells severely decreased, reduced lytic function IL15Ra IL2/15Rb NK cells severely decreased NK cells absent c-kit NK cells decreased, impaired lytic function Reference Wu, 2001 Iizuka, 1999 Smyth, 1999 Ito, 1999 Lodolce, 1998 Gilmour, 2001 Suzuki, 1997 Colucci, 2000 Knockouts/Transgenics: Cytokines Gene Deleted IL15 Flt3-ligand Effect NK cells absent; no lytic function NK cells severely decreased, impaired lytic function Reference Puzanov, 1996 Kennedey, 2000 McKenna, 2000 NK Cell Differentiation Pathway: Informative Gene Knockout and Transgenic Mice HSC >> CLP >> T/NKP >> NKP >> NK Trnscrptn. Fctr./DBP Ikaros PU.1 (P)* Cytokine/Rcptr. Flt3L Sgnl. Trnsdcr. Jak3 Ets1 Id2 CD3e tg FceR1g tg IRF-1 IRF-2 STAT5a/b (P) MEF (P) IL15 IL15Ra IL2/IL15Rb LTa/LTbR (P) III. NK Cell Function as Anti-tumor and Anti-Viral Effector Cells • NK cells were initially described as being cells important for surveillance against tumor development, or more importantly, against tumor metastases • NK cells were also found to be important as anti-viral effector cells, particularly against Herpes virus infection. NK cells and Anti-tumor Activity • • What is the evidence of NK cell anti-tumor function? In vitro – many tumor cells are susceptible to lysis by NK cells depending upon how you assess killing (Kashii, Y., et al. J. Immunol. 163:5358-66 [1999]). In vivo….. Putative Evidence for Immunosurveillance by NK Cells Using Transplantable Tumor Models • • • • Elimination of NK cells resulted in increased tumor growth Elimination of NK cells resulted in increased numbers of metastastic lesions in lungs Adoptive transfer of NK cells, into immunodeficient animals challenged with tumors, results in tumor clearance in metastases models Best results almost always were derived in models of metastatic disease (Barlozzari, T., et al., J. Immunol. 134:2783-2789, 1985) This evidence did not initially garner robust support for NK cell participation in immune surveillance – Why? • • • There has been a growing belief that transplantable tumor models have little value in assessing tumor immunity, and particularly for “immune surveillance” of tumors The only report providing evidence for disease in individuals with reduced NK cells is for recurrent Herpes virus infections (Biron, C.A., et al., NEJM 322:1731-1735, 1989) Identification of receptors on NK cells with coordinate tumor cell ligand was lacking Studies Supporting Increased Incidence of Cancer in Immunosuppressed Individuals • An 11 year follow-up study of immune function and cancer incidence in a general population of 3625 individuals was carried out (Imai, K., et al., The Lancet 356:1795-1799, 2000) • Immune function, i.e. NK cell lytic activity, was assessed at baseline and cancer incidence • Medium and high cytolytic function was associated with reduced cancer risk; low cytolytic function was associated with increased cancer risk Support for NK Cells Providing a Mechanism for Immune Surveillance of Cancer • • • • • Families of NK cell receptors (e.g. NKG2s) with activating and inhibitory function have been defined Tumor associated ligands similar to MHC Class I have been defined, e.g. Rae-1 [mice], MICA/B [humans] Binding of MHC Class I and Class I-related proteins (e.g. Rae-1(a-e); ULBP-1, -2, -3; H60) by NKR has been demonstrated In mice, binding of NKG2D to Rae1a (Cerwenka, A., et al., PNAS USA 98:11521-11526, 2001) or Rae1b (Diefenbach, A., et al., Nature 413:165-171, 2001) has been demonstrated to activate anti-tumor lytic function Human orthologs of Rae-1 genes, e.g. ULBP-1 also are bound by NKG2D; and this activates NK lytic function NK Cells as Anti-viral Effector Cells: Evidence for a role as anti-virus effector cells • Natural defects in NK cells Recurrent Herpes virus infections [Biron, 1989] • Expansion of NK cells during viral infections LCMV infections • Viral antigens as ligands for NK cell receptors ULBP1-4 NK-mediated Response to Virus Infection NK Cells as Anti-viral Effector Cells: Mechanisms of Evasion of NK Cell Function by Viruses • Expression of virally encoded MHC class I protein homologs • Selective modulation of MHC Class I expression by viral proteins • Virus-mediated inhibition of activating receptor function • Production of virally encoded cytokine-binding proteins or cytokine-receptor agonists • Direct viral effects on NK cells – infection/envelope ligation of inhibitory receptors Virus-infected Cell NK Cell MHC ClassI Homolog Selective Expression Activating Receptor Antogonist Virus Down Regulating Activating Ligand MHC Class I Inhibitory Receptor 1 MHC Class I Inhibitory Receptor 2 Activating Receptor 3 Cytokine Binding Protein Cytokine Receptor Cytokine Antogonist 4 Cytokine Receptor NK Cell Infection 5 IV. NK Cell Recognition Receptors • • • • • “Missing Self” Hypothesis Activation and Inhibition via Receptors Recognition of “Self” Recognition of Tumor Cells Recognition of Virus-infected Cells “Missing Self” Hypothesis • • • NK cells do not require expression of MHC Class I determinants for recognition of target cells. There is, in fact, an inverse relationship between expression of MHC Class I and susceptibility to lysis by NK cells, i.e. less Class I equals more lysis. Led to the hypothesis* that NK cells surveyed the surface of target cells for “self”. If it was present, the cell was presumed to be normal and not lysed. If self was absent, as is often the case in tumor cells and virus-infected cells, NK cells could be activated to lyse the “abnormal” cell. *Ljunggren, H.G. and K. Karre, 1990. Immunology Today 11:237-244. Receptors in Innate and Adaptive Immunity Characteristics Innate Adaptive Specificity inherited in the genome Yes No Expressed by all cells of a particular type Yes No Trigger immediate response Yes No Recognize broad classes of pathogens Yes No Encoded in multiple gene segments No Yes Require gene rearrangement No Yes Clonal distribution No No Yes Yes Able to recognize a wide variety of molecular structures Recognition – NK cells - - - There is no evidence supporting clonally restricted recognition molecules expressed by NK cells, nor for recombinatorial events being important for development of an NK cell repertoire NK cells recognize MHC determinants, but these structures, nor peptides expressed by MHC, are target antigens for activation of NK lytic function Some NK cells express CD8 homodimers, but it is unclear whether binding to MHC Class I affects activation NK cell recognition of targets involves a balance between inhibitory signals and activation signals Receptor:ligand pairs providing inhibitory signals are fairly well defined Receptor:ligand pairs providing activation signals are rapidly being defined NK Cell Gene Complex (NKC) • The NKC is a genomic region, first described on NK cells, encoding structurally related receptors • NKC maps to Chromosome 12p13, 6 and 4 in man, mouse and rat, respectively • Type II integral membrane proteins with external domain similar to Ctype (Ca++-dependent) lectins. However, they lack amino acid residues that coordinate binding of Ca++, and do not bind carbohydrates in the same manner as conventional C-type lectins. Can be expressed homoor heterodimers. • Highly conserved evolutionarily – found in sea squirt and several poxviruses • Activating and inhibitory receptors for immune cells; can be either primary or co-stimulatory receptors. NK Cell Gene Complex (NKC) - Contains genes encoding C type lectin related receptors - Disease resistance elements mapped to this locus, e.g. Cmv1 - Conserved across species Human – Chromosome 12 Mouse – Chromosome 6 Rat – Chromosome 4 Leukocyte Receptor Cluster (LRC) LRC is a ~1 mb region located on chromosome 19q13.42 NK Cell Inhibitory Receptors: CLRR and KIR Name p58.1 p58.2 p70 p140 p49 LIR1 LIR2 CD94* NKG2A NKR-P1B, D p40 IRC1 p75AIRM1 Alternative Name[s] KIR2DL1 KIR2DL2 KIR3DL1 KIR3DL3 KIR2DL4 ILT2/LILRB1 ILT4/LILRB2 KLRD1 KLRC1/CD159A CD161B, D LAIR1 IRp60/CMRF35H Siglec-7 *CD94 forms heterodimers with NKG2A, -C and –E **CD94/CD159A heterodimer is specific for HLA-E Cellular Ligand HLA-Cw2,4,5,6 HLA-Cw1,3,7,8 HLA-Bw4 HLA-A3, -A11 HLA-G HLA-G HLA-F HLA-E** HLA-E Clrb ? ? Sialylated sugars Viral Ligand HCMV-UL18 Target Cell membrane NH3 IgV IRp60 NK Cell membrane Cytoplasm SHP-1 COOH I/VxYxxL Inhibition of lytic function ITIM • Immunoreceptor tyrosine-based inhibitory motif • Based upon the amino acid motif: I/VxYxxL • Commonly expressed in signaling receptors in lymphocytes • Recruits SHP-1/SHP-2 phosphatases • Linked to inhibition of function in lymphocytes NK Cell Activating Receptors Name NKp46 NKp30 NKp44 2B4 NTB-A NKp80 CD16 CD2 DNAM-1 NKG2D NKR-P1A NKR-P1C NKR-P1F P40 IRC1 p75AIRM1 Alternative Name[s] Ly94/NCR1 IC7/NCR3 Ly95/NCR2 CD244 KALI KLRF1 FcgRIII LFA-2 CD226 D12S2489E/CD159D CD161A CD161C CD161F LAIR1 IRp60/CMRF35H Siglec-7 Cellular Ligand Viral Ligand ? SV-HA, IV-HA ? ? SV-HA, IV-HA CD48 ? ? IgG CD58, LFA-3 PVR/CD155, Nectin-2/CD112 MICA, MICB, MULT1 ULBP1-4 [IC-21]* ? Clrg ? ? Sialylated sugars *Rat NKR-P1A binds an undefined determinant on IC-21 tumor cells NK cell activating receptors • • • • • • Loss of the inhibitory signal does not, in and of itself, provide signals to kill target cells Some receptors able to activate NK cells to kill target cells have been defined – NKG2D, Ly49D, Ly49H, NKp30, NKp44, NKp46, CD161A Some activating receptors are members of the C-type lectin [e.g. NKG2D] and IgSF [NKp30] superfamilies IgSF members often referred to as KARs Associate with an adaptor molecule [e.g. DAP12] containing an ITAM. Associate via a charged residue in the TM domain Some ligands for activating receptors have been defined, e.g. RAE-1 for NKG2D NH3 NKp46:SV-HA or IV-HA IgC2 IgC2 FceR1g CD3z R NK Cell membrane *D COOH I T A M I T A M I T A M Cytoplasm ZAP70 SYK I T A M Activation NKp44:SV-HA or IV-HA NH3 IgV DAP12 K NK Cell membrane *D Cytoplasm I T A M I T A M ZAP70 SYK COOH Activation NKp30:? [iDCs and some tumors] NH3 IgV CD3z R * COOH NK Cell membrane D I T A M I T A M I T A M I T A M I T A M I T A M Cytoplasm ZAP70 SYK Activation ITAM • Immunoreceptor tyrosine-based activating motif • Based upon the amino acid motif: …YxxL/Ix6-8YxxL/I… • Serves as a signaling partner to transmembrane receptors with a charged residue in the transmembrane region which allows docking of signal transducers such as DAP12, CD3z-CD3z homodimers, CD3zFcer1g heterodimers • Activation of cells either via PI3 kinase, or ZAP70 or Syk tyrosine kinases NKG2D • • • • Single gene Distantly related to other NKG2 family members Alternatively spliced isoforms (short and long) in mice NKG2D-s and NKG2D-l, short from binds both DAP10 and DAP12 • Expressed in NK cells, CD8+ cells and macrophages NKG2D:MICA, MICB, ULBPs COOH COOH CTLD CTLD DAP10 R NK Cell membrane *D Y x x M Cytoplasm PI3K Grb2 NH3 NH3 ERK1/2 MAPK Cytokine secretion Cytotoxicity Ligands for NK Cell Activating Receptors • • • • • • MICA, MICB: Stress-inducible molecules encoded within the human MHC, also can be induced by some infections. Normally expressed by gastrointestinal epithelium, but also by some epithelial, lung, breast, kidney, ovary, prostate and colon tumors, and by some melanomas. Transmembrane with a1, a2, and a3 domains; but do not associate with b2m and do not bind peptides. ULBP1-4: 1-3 are GPI-linked, cell surface molecules which bind human cytomegalovirus UL-16; ULBP-4 is a cell surface molecule with transmembrane and cytoplasmic domains. ULBPs have a1 and a2 MHC Class I-like domains. Rae1b: Retinoic acid inducible protein, in mice, that shares sequence homology with ULBPs. Expressed in early embryogenesis and in some tumors, but generally absent in normal tissues. H60: Minor histocompatibility antigen expressed by Balb/c mice, target for alloreactivity responses by C57Bl/6 mice. DCs: Known that NKp30 is required for recognition of immature DCs by activated NK cells. IC-21: Known that rat CD161A is required for recognition of IC-21 tumor cells to mediate their lysis. Signal Transduction Pathway for NK cells* (NKp44) (KIR2DL1) (NKG2D) (DAP12) Modified from Vely and Vivier, 2005, www.stke.org/cgi/content/full/sigtrans;2005/292/cm6 V. Non-adaptive vs. Adaptive Function - Mediators of non-adaptive immunity - Interface between non-adaptive and adaptive immunity – “Passive” interaction – antibody dependent cellular cytotoxicity “Active” interaction – reciprocal co-activation of NK cells and DCs to induce adaptive responses - New hypothesis regarding NK cells as mediators of adaptive immunity is the topic of the journal club article Interactions with Dendritic Cells to Promote Adaptive Immune Responses VI. Therapeutic Applications of NK Cells Biological Response Modifiers IL2, IL12, IL15, IL21, IFNa, IFNg, IFNb, PolyI:C, b-glucan Adoptive Cellular Immunotherapy Freshly isolated NK cells – autologous/allogeneic-alloreactive BRM/Cytokine activated NK cells – autologous/allogeneic Long term established NK cell lines (NK-92) NK Cells as Vehicles for Delivery of Therapeutic Agents Chemotherapeutic agents - doxorubicin Cytokines – IL2 Trials for: melanoma, renal cell carcinoma, lung carcinoma, ovarian cancer, Glioblastoma – variable results Utilization of Modified NK-92 Modification of functional activity IL2 Prolonged in vivo activity NK-92 Control of in vivo Expansion NK-92-CD20 Targeting specific tumor types NK-92-Her2/neu NK-92-CD19 NK-92-CD38 Epithelial tumors breast ovarian Myeloma In vivo control of proliferation through suicide gene binding B-cell precursor leukemia Improved cytolytic efficacy Accessibility to resistant tumors Modified from Suck, G. 2006