* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Population Genetics 2: Linkage disequilibrium Consider two loci and
Nutriepigenomics wikipedia , lookup
Gene desert wikipedia , lookup
Inbreeding avoidance wikipedia , lookup
Ridge (biology) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Genome evolution wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Koinophilia wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene expression profiling wikipedia , lookup
Human genetic variation wikipedia , lookup
X-inactivation wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genome (book) wikipedia , lookup
Genetic drift wikipedia , lookup
Gene expression programming wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Population genetics wikipedia , lookup
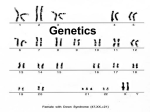
Population Genetics 2: Linkage disequilibrium Consider two loci and 1 generation of random mating: A gene: AA, Aa, and aa B gene: BB, Bb, and bb Genotype frequencies in a population A gene B gene 2 2 fAA = p fAa = 2pq 2 faa = q fBB = x fBb = 2xy 2 fbb = y p+q=1 x+y=1 Random association of alleles at a single locus: HWE What about random association of alleles at different loci after random mating? 1 Consider two loci and 1 generation of random mating: Random association in gametes Alleles at B locus Alleles at A locus A(p) a(q) B (x) AB (px) aB (qx) b (y) Ab (py) ab (qy) remember: p + q =1 and x + y = 1 Random association of alleles at a different locus: LINKAGE EQUILIBRIUM GAMETIC PHASE EQUILIBRIUM Consider two loci and 1 generation of random mating: Surprisingly common result: Gene A: HWE Gene B: HWE Gene A + Gene B: disequilibrium 2 Consider two loci (on different chromosomes) and 1 generation of random mating: Example: Population 1: 100% AABB Population 2: 100% aabb Mix populations equally: 50% AABB + 50% aabb 1 generation of random mating (only three matings possible) : AABB x AABB = AABB aabb x aabb = aabb AABB x aabb = AaBb Nine genotypes are possible: AaBB aaBB aaBb AABb AAbb Aabb AABB aabb AaBb We only see 1/3 after 1 generation of random mating! They did not reach equilibrium after one generation of random mating. With continued random mating the “missing” genotypes would appear, but not immediately at their equilibrium frequencies! Consider two loci and 1 generation of random mating: - attainment of linkage equilibrium is gradual - about 50% of disequilibrium “breaks down” per generation - linkage disequilibrium (LD) persists in populations for many generations - LD = gametic phase disequilibrium 3 LD in individuals (BIOL 2030 stuff): We need a new symbolism Case 1: AB gamete + ab gamete = AaBb Case 2: Ab gamete + aB gamete = AaBb New symbolism: AB/ab indicates the union of AB gamete + ab gamete LD in individuals: Let’s take an AB/ab individual as an example: Physical linkage: A B What types of gametes can the AB/ab make? a b By the way, lets assume physical linkage. (1) AB (2) ab (3) Ab (4) aB Notation = AB/ab Parental or non-recombinant gametes Non-parental or recombinant gametes 4 Review meiosis in a single meiocyte: Gametes Notation: Parental (non-recombinant): A B Parental configuration Recombinant: a B Recombinant: A b Parental (non-recombinant): a b 1 crossing over event: 50% parental and 50% recombinant !!! LD in individuals: When genes are on the same chromosome: Physical linkage: A B fAB = fab ≥ fAb = faB a b f (non-recombinant) ≥ f (recombinant) Notation = AB/ab Recombination fraction (r) is the proportion of recombinant gametes produced by an individual. When r = 0: fAB + fab = 100% [fAb + faB = 0%] When r = 0.5: fAB + fab = 50% [fAb + faB = 50%] 5 From: iGenetics P. J. Russell (2002) page 350 LD in individuals: When genes are on different chromosomes: Un linked genes: A fAB = fab = fAb = faB f (non-recombinant) = f (recombinant) a B b r = 0.5 • when genes are on different chromosomes • when genes are on same chromosome and recombination is high enough for independent assortment 6 LD in individuals: Individual AB/ab produces the following: (1) AB: fAB = 0.38 (2) ab: fab = 0.38 (3) Ab: fAb = 0.12 (4) aB: faB = 0.12 r = 0.12 + 0.12 = 0.24 What do we expect for individuals if allelic association is random? What do we expect in a population if allelic association is random? LD in populations: Random association in gametes Alleles at B locus Alleles at A locus A(p) a(q) B (x) AB (px) aB (qx) b (y) Ab (py) ab (qy) remember: p + q =1 and x + y = 1 fAB =px fab = qy fAb = py faB =qx fAB + fab + fAb + faB = 1 7 LD in populations: The frequency of an AB gamete in a population has two sources: A B AB • Some individuals have this configuration: non-recombinant [parental] • Some individuals produce this as a recombinant configuration. LD in populations: f AB ' - recombinants recombinants non = (1 − r ) f AB + ( r) px frequency of AB probability of no recombination gametes in last generation prob of prob of putting recomb together A and B at random from recombinants 8 ' f AB = - recombinants recombinants non (1 − r ) f AB + ( r) px frequency of AB probability of no recombination gametes in last generation Obs. – exp. prob of prob of putting recomb together A and B at random from recombinants ' f AB − px = (1 − r )( f AB − px ) D = (1 − r )( f AB − px) = the linkage disequilibrium parameter fAB = px + D In excess due to LD fab = qy + D fAb = py - D faB =qx - D Deficient due to LD Remember: Individual AB/ab produces the following: (1) AB: fAB = 0.38 (2) ab: fab = 0.38 (3) Ab: fAb = 0.12 (4) aB: faB = 0.12 r = 0.12 + 0.12 = 0.24 What do we expect for an individuals if association is random? What do we expect in a population if association is random? 9 Forces that increase D in populations: 1. Migration 2. Natural selection 3. Genetic Drift LD in populations: • Comparing D among populations is difficult. • Standardize D as a fraction of the theoretical maximum for the popn D Dmax D = f AB × f ab − f Ab × f aB excess due to LD deficient due to LD Dmax = qx or py (whichever is smaller) 10 LD in a population: Example: blood group polymorphism in a sample of 1000 British people MN blood group Gamete frequencies expected fM = p = 0.5425 MS = 474/2000 = 0.2370 (+) px = 0.1671 fn = q = 0.4575 Ms = 611/2000 = 0.3055 (-) py = 0.3751 nS = 142/2000 = 0.0710 (-) qx = 0.1409 ns = 773/2000 = 0.3865 (+) qy = 0.3166 Ss blood group D = ef × ef × ef MS ns − ef Ms nS non recombinant recombinant Dmax = qx or py € fS = x = 0.3080 fs = y = 0.6920 D = (0.2370)(0.3865) – (0.3055)(0.0710) = 0.07 Dmax: qx = 0.14 or py = 0.37; so Dmax = 0.14 D is (0.07/0.14)*100 = 50% of the theoretical maximum Homework: Genotype counts in the population Gametes MN locus Ss locus MS = 474 MM = 298 MN = 489 NN = 213 SS = 483 Ss = 418 ss = 99 NS = 142 Ms = 611 Ns = 773 Use chi-square test to: 1. determine if each locus is in HWE 2. determine if gamete frequencies are in equilibrium 11 Recombination reduces LD: Rate of decay of LD under various recombination rates Standardized disequilibrium D/Dmax 1 0.9 r= 0.001 0.8 0.7 0.6 r= 0.01 0.5 0.4 0.3 0.2 r= 0.1 0.1 r= 0.5 0 1 9 17 25 33 41 49 57 65 73 81 89 97 generations Recombination reduces LD: “Hitchhiking” of a mutator gene with and without recombination No recombination r=0 Recombination r = 0.5 Mutator allele that increase the mutation rate Beneficial allele subject to strong positive selection Adapted from Sniegowski et al. (2000) BioEssays 22:1057-1066. 12 Mapping disease genes: 1. Family studies • Uses family pedigrees • Co-segregation of disease and a marker on the pedigree 2. Allelic association studies (LD mapping) • Uses population data • Relies on strong LD only among closely linked loci • Sample affected and unaffected individuals • Very large samples are required! • Look for markers with more LD in affected individuals Both approached have powers and pitfalls Mapping disease genes: r = 0.01 (1 centiMorgan) is about 1million bp in humans 13 female-limited Batesian mimicry 5-locus linkage group (supergene): § Tail length § Hind-wing pattern § Forewing pattern § Epaulet color § Body color Selection: Bitter tasting Only certain complex color morphs provide gains in fitness Tasty mimics (Papilio memnon females) LD: Few maladaptive patterns produced per generation due to linkage male Class I Class III Class II The MHC locus on human chromosome 6 LD over 10s-100s million of bp due to selection for combinations of loci 14 Sexual reproduction reduces LD Linage disequilibrium: Keynotes • Attainment of equilibrium at different loci is gradual; > 1 generation of random mating. • Physical linkage slows the rate to equilibrium even more! • “r” determines the rate to equilibrium, the lower the fraction, the longer to equilibrium. • When r = 0.5 the loci are said to be un-linked; such loci are very far apart on the same chromosome, or in different chromosomes. When r < 0.5 the genes are said to be linked. When r =0 the loci are in permanent disequilibrium. • Disequilibrium can arise from sources other than linkage: o o o o • Admixture of populations Natural selection acting on one or more of the loci Inbreeding in plants that regularly undergo self-fertilization Genes located in a chromosomal inversion (SUPERGENE) The term LINKAGE DISEQUILIBRIUM is used to describe any source of disequilibrium, regardless of whether the two genes are physically linked or not. 15