* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Virtual Screening in Drug Discovery: an Overview
Toxicodynamics wikipedia , lookup
CCR5 receptor antagonist wikipedia , lookup
Discovery and development of tubulin inhibitors wikipedia , lookup
Compounding wikipedia , lookup
Discovery and development of neuraminidase inhibitors wikipedia , lookup
Nicotinic agonist wikipedia , lookup
Discovery and development of antiandrogens wikipedia , lookup
NK1 receptor antagonist wikipedia , lookup
Discovery and development of ACE inhibitors wikipedia , lookup
Discovery and development of non-nucleoside reverse-transcriptase inhibitors wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Discovery and development of direct Xa inhibitors wikipedia , lookup
Theralizumab wikipedia , lookup
Neuropsychopharmacology wikipedia , lookup
Pharmacognosy wikipedia , lookup
Prescription drug prices in the United States wikipedia , lookup
Prescription costs wikipedia , lookup
Drug interaction wikipedia , lookup
DNA-encoded chemical library wikipedia , lookup
Pharmaceutical industry wikipedia , lookup
Neuropharmacology wikipedia , lookup
Pharmacokinetics wikipedia , lookup
Discovery and development of integrase inhibitors wikipedia , lookup
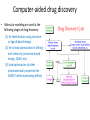
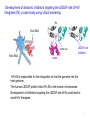
Molecular Modeling in Drug Discovery: an Overview Nanjie Deng Drug development is a long and expensive process Early stage drug discovery Pharmacokinetic Preclinical safety Phase I, II, III Target identification Hit /Lead identification Lead optimization Preclinical development Clinical development • It takes > 10 years and > 1 billion dollars of investment to develop a new drug •Molecular modeling tools has been widely used in drug development projects to help reduce the time/cost in early stages of drug discovery. The goal of early stage drug discovery: to identify hits and leads compounds • Most drugs work by interacting with a protein/DNA at a specific target site. • Drug molecules need to (1) bind tightly to the target site (affinity) and exert the desired activity (potency) (2) have minimal off-target binding (side effects/toxicity) (3 ) can get to the target site and can be removed from the body by metabolism and excretion pathways. (ADME properties) • Three key steps in early stages of drug discovery: Discover hits: molecules showing some activity. Discover leads: a series of related molecules that show some variation in activities as the structure is modified. Optimize Leads: further optimization resulting in drug candidates with the desired potency and good ADME/T properties. Computer-aided drug discovery • Molecular modeling are used at the following stages of drug discovery (1) hit identification using structureor ligand-based design. (2) hit-to-lead optimization of affinity and selectivity (structure-based design, QSAR, etc.) (3) lead optimization of other pharmaceutical properties like ADMET while maintaining affinity Two main approaches to computer-aided drug design • Ligand-based relies on knowledge of known molecules that bind to the biological target. These existing molecules are used to derive a pharmacophore model that defines the minimum necessary structural characteristics a molecule must possess in order to bind to the target. • Structure-based - uses the knowledge of the 3D-structure of the target to design and predict candidate drugs that bind with high affinity and selectivity to the target. Methods used in structure-based drug design • Virtual screening identification of new ligands for a given receptor by searching large databases of small molecules to find those fitting the binding pocket of the receptor using fast approximate docking programs. • De novo ligand design ligand molecules are built up within the binding pocket by assembling small molecular fragments. The method allows novel structures to be designed. Development of allosteric inhibitors targeting the LEDGF site of HIV Integrase (IN): a case study using virtual screening IN Viral DNA LEDGF site Host DNA LEDGF-site inhibitor LEDGF HIV-IN is responsible for the integration of viral the genome into the host genome. The human LEDGF protein links HIV-IN to the human chromosome. Development of inhibitors targeting the LEDGF-site of IN could lead to novel HIV therapies. 7 Discovery of HIV-1 Integrase inhibitors: a virtual screening workflow A commercial library of 200,000 compounds 160,000 compounds 200 molecules Generation of pharmacophore 25 molecules purchased for testing Compound 1 was found to inhibit IN/LEDGF binding at 100 mM. Christ et al., Nature. Med. Chem., 2010