* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Group selection wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Point mutation wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Heritability of IQ wikipedia , lookup
Designer baby wikipedia , lookup
Inbreeding avoidance wikipedia , lookup
Gene expression programming wikipedia , lookup
Genetics and archaeogenetics of South Asia wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Koinophilia wikipedia , lookup
Human genetic variation wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Genetic drift wikipedia , lookup
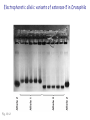
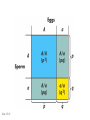
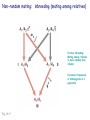
Chapter 19: Population genetics Fig. 19-1 Population genetics Population: interbreeding members of a species Three major principles of Darwinian evolutionary theory: • variation for traits exists within populations • selection applies to a subset of those traits (selection can act only upon variations) • traits are genetically transmitted Polymorphism: multiple forms of a gene are commonly found in a population (all studied populations are “wildly” polymorphic) • chromosomal polymorphisms • immunological polymorphisms • protein polymorphisms • nucleic acid sequence/structure polymorphisms p = (2 x MM) + (1 x MN) = frequency of M allele q = (2 x NN) + (1 x MN) = frequency of N allele Therefore, frequency of MN reflects the genetic variation in a population Fig. 19-2 “Factoids” regarding protein polymorphisms in most species: • structural polymorphisms are displayed by about one-third of all proteins • typically, about 10% of the individuals in a large population are heterozygous for polymorphisms of an average gene Therefore, enormous protein-level variation exists in most populations Electrophoretic allelic variants of esterase-5 in Drosophila Fig. 19-2 Electrophoretic variants of hemoglobin A in humans Fig. 19-3 Enormous naturally-occuring variation (polymorphism) in protein sequence Enormous naturally-occuring variation (polymorphism) in chromosomal rearrangements Restriction sites within Drosophila xdh gene (58 wild chromosomes sampled) 4-base sites in 4.5 kb DNA * Site present in minority of chromosomes 0/1 Site in ½ of chromosomes Fig. 19-5 Enormous naturally-occuring variation (polymorphism) in nucleotide sequence Enormous naturally-occuring variation (polymorphism) in tandem repeat arrays (VNTRs) Hardy-Weinberg Equilibrium Random mating within a large population assures a stable equilibrium of genetic diversity in subsequent generations provided that certain assumptions apply: Mating is random (no biased mating, infinite population size) Allele frequencies do not change (no selection, no migration, etc.) Hardy-Weinberg Equilibrium For a two-allele system, all genotypes exist as a simple product of the frequency of each allele: homozygotes = p2 or q2 heterozygotes = 2pq p2 + 2pq + q2 = 1 Box 19-2 Box 19-2 Allele frequencies determine frequencies of homozygotes and heterozygotes Fig. 19-6 Rare alleles are almost always found in heterozygotes, almost never homozygous Another measure of heterozygosity is haplotype diversity Haplotype: combination of non-allelic alleles on a single chromosome MN allele and genotype frequencies reflect Hardy-Weinberg assumptions Allele and genotype frequencies can vary between populations, while exhibiting H-W equilibria within each population Non-random mating: inbreeding (mating among relatives) Positive inbreeding: Mating among relatives is more common than random Increases frequencies of homozygotes in a population Fig. 19-7 Extreme inbreeding: self-fertilization results in loss of heterozygosity No change in p or q; change only in heterozygosity and diversity Fig. 19-8 Negative inbreeding (enforced outbreeding) -barriers to inbreeding are common attributes of successful populations Positive assortative mating - individuals chose “like” mates (not necessarily relatives) Negative assortative mating - individuals choose dissimilar mates Sources of variation • Mutation – very slow • Mutation is the ultimate source of variation But spontaneous mutations occur at extremely low frequencies Mutation frequency is influenced by allele frequency Mutation alone is a very slow evolutionary force and cannot directly account for diversity observed in populations. Box 19-3 Sources of variation • Mutation – very slow • Recombination – rapidly mixes genes to provide new genetic combinations in a population • Migration – gene flow among different populations changes gene frequencies Selection: directed change in genotypes in a population Fitness: survival and reproduction success; function of genotype and environment Fitness can be obvious (mortality, sterililty) • HbS/HbS: severe anemia, low survival • HbS/HbA: apparent resistance to malaria or more subtle/partial/conditional Fitness (viability) of various homozygotes as a function of temperature Drosophila pseudoobscura Fig. 19-9 Enhanced fitness of a genotype will enrich those genes in subsequent generations of that population Frequencies of positively selected genes increase over time Frequencies of negatively selected genes decrease over time Change in A frequency (p) is greatest where p = q Fig. 19-11 For a two-allele system, mean fitness (W) in a population is the proportional contribution of fitness by each genotype (A/A, A/a, a/a) W = p2WA/A + 2pqWA/a + q2Wa/a WA/A and WA/a > Wa/a p should increase q should decrease Wa/a > WA/A and WA/a q should increase p should decrease Fitness can account for allele frequency changes over time Fig. 19-12 p for malic dehydrogenase electrophoretic mobility variant MDHF where WS/S=1, WS/F=0.75, WF/F=0.4 Selection: directed change in genotypes in a population Fitness: survival and reproduction success; function of genotype and environment Frequency independent selection: fitness is independent of genotype frequency Frequency dependent selection: fitness changes as genotype frequency changes Random genetic drift: random changes in gene frequency that can lead to extinction/fixation of genes • Requires no selection • Essentially “sampling error” inherent in each generation in achieving HardyWeinberg equilibrium • Most exaggerated in small populations (especially “founder effects”) • Allows isolated populations to diverge without differential selection (each experiences its own drift history) Model: history of emergence of ten mutations and their drift in a population over time Fig. 19-13 Drift to extinction for nine; drift to p=1 in one Drift explains differences in unselected allele frequencies in isolated populations Genetic change is directed by diverse evolutionary forces which tend to increase (blue) or decrease (red) variation Recommended problems in Chapter 19: 3, 5, 12, 17