* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Signal Transduction in Bacterial Chemotaxis
Hedgehog signaling pathway wikipedia , lookup
Protein moonlighting wikipedia , lookup
Phosphorylation wikipedia , lookup
Protein phosphorylation wikipedia , lookup
List of types of proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Biochemical cascade wikipedia , lookup
Paracrine signalling wikipedia , lookup
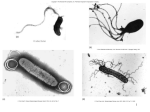
Victor Sourjik Signal Transduction in Bacterial Chemotaxis Introduction Chemotaxis in Escherichia coli is one of the moststudied model systems for signal transduction. E. coli can respond to a variety of amino acids, sugars, and dipeptides, as well as pH, temperature, and redox state, by adjusting its swimming behaviour. The signalling pathway in chemotaxis is relatively simple (Fig. 1) but noted for its high sensitivity, integration of multiple stimuli, wide dynamic range, and robustness. Signalling involves stimulus-dependent autophosphorylation of the histidine kinase CheA and subsequent phosphorylation of the response regulator CheY. The pathway also includes CheZ, a phosphatase of CheY-P, and an adaptation system that consists of the methyltransferase, CheR, and the methylesterase, CheB. Our goal is a quantitative description of signal processing in chemotaxis. We apply several fluorescence microscopy techniques to study dynamics of the pathway in the cell. We also investigate cross-talk between chemotaxis and other cellular networks and study effects of the natural cell-tocell variation in protein levels on signal transduction. In collaboration with computer modelling groups, we integrate our results with available biochemical data to reproduce pathway behaviour in silico. Fig. 1: Signal transduction pathway in chemotaxis of E. coli FRET-based measurement of signal processing D. Kentner, S. Schulmeister We developed an assay, based on fluorescence resonance energy transfer (FRET), to study in vivo interactions of E. coli chemotaxis proteins expressed as fusions to cyan fluorescent protein, CFP, and yellow fluorescent protein, YFP (Fig. 2). So far, we used FRET to probe phosphorylation-dependent interactions of the response regulator CheY with CheZ, CheA, and the motor protein FliM. We are now extending these studies to map all pairwise interactions between chemotaxis proteins. FRET enables us to measure the amplitudes and kinetics of changes in interaction upon stimulation and to estimate protein binding constants in vivo. We use FRET between CheY and CheZ as a reporter assay to monitor output of the receptorCheW-CheA sensory complex and to quantify amplification and integration of different stimuli. Another part of the project is to analyze the amaz- Victor Sourjik [email protected] 1997 PhD (Dr. rer. nat.) at the University of Regensburg 1998-2003 Postdoctoral work at the Department of Molecular and Cellular Biology, Harvard University, USA since 2003 Group leader at the ZMBH 108 ZMBHReport04.indd 108 10/05/05 9:41:48 Fig. 2: FRET as a reporter of pathway activity. Stimulation-induced changes in FRET between CheY-YFP and CheZ-CFP are shown on the right. ing dynamic range of the chemotaxis system - its ability to respond and perfectly adapt to over five orders of magnitude of chemoeffector concentrations. Formation and dynamics of chemoreceptor cluster S. Röhrig Sensory complexes are organized in polar receptor clusters that are essential for signal processing and include thousands of proteins (Fig. 3). The goal of this project is to investigate the assembly and positioning of the receptor cluster. We use time-lapse fluorescence microscopy to study the formation of the new clusters during cell division, and fluorescence recovery after photobleaching (FRAP) to measure association dynamics of the proteins that constitute the sensory complex. External funding During a part of the period reported our research was supported by a project grant (SO 421/3-1) from the Deutsche Forschungsgemeinschaft. PUBLICATIONS 2002 - 2004 Sourjik, V. and Berg, H. C. (2002a). Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 99, 123127. Sourjik, V. and Berg, H. C. (2002b). Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 99, 12669-12674. Sourjik, V. and Berg, H. C. (2004). Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437441. Liberman, L., Berg, H. C., and Sourjik, V. (2004). Effect of chemoreceptor modification on assembly and activity of the receptor-kinase complex in Escherichia coli. J. Bacteriol. 186, 6643-6646. Sourjik, V. (2004). Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 12, 569-576. THESES Diploma Løvdok, Linda (2004): Cell-to-cell variation in gene expression in Escherichia coli and its effect on signal transduction in chemotaxis. Contact: ZMBH, Im Neuenheimer Feld 282 69120 Heidelberg, Germany Fig. 3: Chemoreceptor clustering in bacteria. Overlay of receptor clusters (labeled by CheR-YFP; green) and cell division ring (labeled by FtsZ-CFP; blue) with a bright-field image of the cell. Phone: +49 6221 546858 Fax: +49 6221 545894 [email protected] http://www.zmbh.uni-heidelberg.de/Sourjik/default.html SOURJIK GROUP Group Leader Sourjik, Victor, Dr. PhD Students Kentner, David, Dipl. Biol. Röhrig, Sebastian, Dipl. Biol. Schulmeister, Sonja, Dipl. Biol.* *only part of the time reported Diploma Student Løvdok, Linda* Techn. Assistants Müller, Anette* Schwarz, Gabi Liberman, Louisa* Victor Sourjik 109 ZMBHReport04.indd 109 10/05/05 9:41:52