* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Antibody structure
Duffy antigen system wikipedia , lookup
Innate immune system wikipedia , lookup
Cancer immunotherapy wikipedia , lookup
Complement system wikipedia , lookup
Monoclonal antibody wikipedia , lookup
Molecular mimicry wikipedia , lookup
Immunosuppressive drug wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
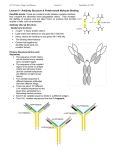
Antibody structure Overview 1. Antibodies belong to a class of proteins called immunoglobulins 2. Antibody molecules belong to one of five classes i.e. IgG, IgM, IgA, IgD & IgE 3. Immunoglobulins are “Y” shaped proteins. The “arms” of the “Y” bind antigens. The tail of the “Y” is responsible for biological activity eg. C’ activity or binding to cells 4. Ability of immunoglobulins to bind antigen determ. by AA sequence in variable region General Characteristics of antibody Antigen-specific products of B cells First of the molecules participating in immune response to be characterized, best understood Basic building block, immunoglobulin domain, is used in molecules of both immune system & other biological recognition systems Antibody molecule has 2 separable functions: 1) specific binding to antigen eliciting response 2) biological activity - recruit cells & components designed to destroy agent to which ab developed Serum and plasma Serum Blood Plasma Blood + anticoagulant Cells Clot Isolation & characterization Chromatography Molecular sieving Ion exhange Affinity Salting out/ Dehydration Ammonium sulfate Alcohol precipitation Ultracentrifugation Immunochemical Chromatography Molecular sieving + + - + + + + + Ion exchange Affinity Salting out Ammonium sulfate (half saturated) Ethanol (90%) Density gradient ultracentrifugation Displacement = Svedberg Kabat & Tiselius (1939) + Albumin a b g Globulins -Discovered that hyperimmunizing rabbits resulted in increased g -Purification revealed antibody activity resided in this serum portion Electrophoretic analysis of serum Sample application Separation by charge - + Anode - + Cathode + Albumin a b Globulins g Protein concentration Protein concentration Characterizing chains Albumin Broad peak of g = heterogenous proteins a1 a2 b + g normal - Need pure Ig to chemically analyze Narrow peak of g = homogenous proteins - + multiple myeloma Bence-Jones proteins in urine of multiple myeloma pts. = dimers of immunoglobulin light chains, k or l Porter (England) Treatment with proteolytic enzyme, papain, resulted in three approximately equal sized fragments: 2 capable of ag rx (fragment antigen binding) 1 could be crystallized (fragment- crystallizable) Fab Fab SS Fab specifically bind antigen, univalent (can’t ppt) one binding site each, identical to each other SS SS Fc Fc crystallizable (thus homogenous) can’t bind ag responsible for biological activity of molecule after ag bound to Fab portions Eddleman (USA) Treatment with mercaptoethanol = 4 chains: (mercaptoethanol breaks S-S bonds) SH HS SH HS SH HS 2 chains = 53,000 daltons 2 chains = 22,000 daltons All immunoglobulins basic unit consisting of 4 polypeptides i.e. 2 H, 2 L Proteolytic enzyme digestion reveals N- & C-terminal N-terminus Fab Papain digestion results in cleavage @ “N” terminus in proline hinge region @ disulfide bridge Fab SS SS SS Fc Fab2 SS Pepsin digestion results in cleavage @ “C” terminal portion, resulting in a divalent fragment (Fab2) joins by S-S & several Fc fragments SS SS C-terminus Cleavage of Ig SH HS SH HS SH HS Fab Papain 4 polypeptide chains Fab SS SS SS 2 Fragment ag binding 1 Fragment crystallizable 1 Fab2 SS SS SS Several small pieces Fc Myeloma cells Plasma cell becomes tumorous = myeloma produces homogenous (monoclonal) Ig Myeloma Ig = myeloma “proteins” Myeloma proteins may be purified & structurally analyzed AA sequence in L & H chains Normal, non immune Normal, immune Myeloid myeloma Light chain sequences N-terminus Fab N-terminus Fab SS SS 214 AA in two domains SS C terminus Analysis from several myelomas C terminus Different sequences in N-termin. domain Variable (VL) Identical sequences in C-termin. domain Constant (CL) Heavy-chain sequence N-terminal domain varies Heavy chain consists of 445 AA in 4 domains (HV) Other 3 domains constant (HC) Flexible Hinge Region Fab N-terminus Fab SS SS SS C terminus % var Sequence variability not distributed evenly in variable region CDR 1 CDR 3 CDR 2 Variation restricted to 3 regions Hypervariable regions (HV1,HV2,HV3) Location of antigen binding site AA sequence det. shape of ag binding site (paratope) Determine epitopes to which Ig binds = complementary- determining regions (CDR) CDR = 6-10 AA CDR 1 = 24 - 34 CDR 2 = 50 - 56 CDR 3 = 89 -97 % var Framework regions = regions where AA seq. is relatively constant (FR) Heavy chain CDR CDR 1 = 31-35 CDR 2 = 50-65 CDR 3 = 95-102 Wu & Kabat plot Structure of variable & constant domains (X-ray crystallography) Light chain C domain Two layers, linked by disulfide bonds Light chain V domain N terminus CDR Disulfide bonds Each layer formed several stretches 3 conformation = b strand Layers = b sheet CDR 1 CDR 2 Order of b strands is characteristic for each sheet 3 D structure = Ig fold C terminus B strands B strands Opening to reveal b strands comprising each b sheet Principal difference between C & V domains V domain has 2 more b strains forming extra loop unique strands C domain V domain Order & orientation characteristic for each domain b strands lettered sequentially according to occurrence in AA sequence in dom. Location of CDR & FR in Ig L & H chains CDR 1 FR1 2 FR2 FR1 3 FR3 FR2 1 FR4 FR4 CH1 FR3 2 CDR CL 3 H L & H chain folding to yield 3 CDR in each chain to form walls of ag binding groove Immunoglobulin consists of 4 polypeptide chains N terminus Ag binding site Ag binding site Variable regions VH VL CH1 CL CH2 Constant region CH3 C terminus Disulfide bonds EM Rabbit Ig X 2,042,500 Light chain Heavy chain Immunologic analysis of immunoglobulins Ig (like most other proteins) stimulate ab in other animal species All species have two major classes of L chains i.e. k,l individual of species produces both types; ratio of k : l varies by species (e.g. mouse 95%k ; human 60% k) in any Ig molecule, both L chains = either k or l, never one of each Ig of all species consist of 5 classes (isotypes) differ in structure of H chains H chains among isotypes differ serologically, CHO content, size & biological function H chains H chain confers unique biologic properties of molecule e.g. 1/2 life, receptor binding , enzyme-, C’ activation with ag Immunoglobulin isotype IgM IgG IgA IgD IgE Heavy chain m g a d e Individual of species produces all H chains, in proportion characteristic for species, but ab molecule H chains are identical ( i.e., 2e, 2d etc.) Isotypic (Class) structure of Ig L H L H L H L H IgG isotype = k2g2 or l2g2, but not k1l1g1a1 IgE isotype = k2e2 or l2e2 ….etc…... H chains H chain confers unique biologic properties of molecule e.g. 1/2 life, receptor binding , enzyme-, C’ activation with ag Immunoglobulin isotype IgM IgG IgA IgD IgE Heavy chain m g a d e Individual of species produces all H chains, in proportion characteristic for species, but ab molecule H chains are identical ( i.e., 2e, 2d etc.) Immunoglobulin consists of 4 polypeptide chains N terminus Ag binding site Ag binding site Variable regions VH VL CH1 CL CH2 Constant region CH3 C terminus Disulfide bonds EM Rabbit Ig X 2,042,500 Light chain Heavy chain Isotypic (Class) structure of Ig L H L H L H L H IgG isotype = k2g2 or l2g2, but not k1l1g1a1 IgE isotype = k2e2 or l2e2 ….etc…... Hinge region Consists of approx 12 AA between CH1 & CH2 No homology between hinge & other Heavy chain domains Angle = 900 0 AA sequence unique for each class Angle = 60 & subclass Angle = 0o AA sequence in hinge region Hinge region Light Chain Cys -Arg-Val-Glu-Pro-Lys-Ser- Cys-Asp-Lys-Thr-His-Thr-Cys-Pro-Pro-Cys -Pro-Ala-Pro-Glu-Arg-Val-Glu-Pro-Lys-Ser- Cys-Asp-Lys-Thr-His-Thr-Cys-Pro-Pro-Cys -Pro-Ala-Pro-GluCys Light Chain Papain Fab region Fc region Characteristics of hinge region Immunoglobulins (with possible exception of IgM & IgE) contain hinge between CH1 & CH2 No homology between AA sequence of hinge & heavy chains AA sequence differs with different classes Comprised of many cysteine and proline residues Cysteine involved in formation of interchain disulfide bonds Proline prevents folding in a globular structure, allowing flexibility between two Fab arms of the Y-shaped antibody; allows open & close to accommodate binding to two epitopes; because it is open, it can be cleaved by proteases (e.g. papain) to generate the Fab & Fc fragments IgG Highest concentration in serum Plays major role in immune dfns. MW approx. 150 kDa Small size ppt in surfaces (e.g. cross placental barrier) Opsinize, aggl. & ppt ag Only activates classical C’ p’way IgG1 Subclasses show close overall relation Heavy chains = g1,g2,g3,g4 IgG2 All normal indiv. have all IgG1>IgG2>IgG3>IgG4 IgG3 has shortest 1/2 life, highest catabolic rate, highest # S-S IgG4 = monoval. no aggl ppt rx., autoab to clot fac Autoab to DNA = IgG1,IgG3 IgG3 IgG4 Structural features of IgG Two g H chains, Two L chains (either k or l but not both) Each H chain = 50 kD Each L chain = 25 kD HV Variable 150 kD, 7S, “g globulin” Least anodic of all serum proteins Constant - + Anode Cathode Structural features of IgM SS SS SS Pentamer (5) First Ig produced following immun. Macroglobulin (M) 900 kD, 19S J chain Doesn’t have hinge region has additional H domain Has a J chain (one of 2 Ig isotypes) 15 kD Structural features of IgA Secretory piece J chain Principal Ig in external secretions e.g. saliva, mucus,sweat, gastric fluid & tears Major Ig of colostrum & milk, provides neonate with major source of intestinal protection against pathogens 165 kD, 7S, migrates as fast g Plasma cell forms basic IgA molecule with J chain which form dimers (second Ig to contain J chain) When released from plasma cell, bind to basal membranes of epithelia via secretory piece In serum, primarily a monomer, no secretory piece Secretory component protects IgA from proteolytic digestion Structural features of IgD Primarily a B cell antigen receptor Long exposed hinge region 170kD, 7S, migrates as fast g No interchain S-S bridges in H chains Readily denatured Structural features of IgE Sometimes called reaginic ag Mediates allergies (Type I hypersensitivies) 190 kDa, 8S, migrates as fast g Contains an extra domain (CH4) which binds to mast cells & basophils May remain attached for long time when ag reappears, cross links IgE on mast cell surface, release mast-cell granules & signs of anaphylaxis Immunoglobulin variations Isotype = class of Ig, all people have Allotype = genetic difference, depends on existence of allelic forms of the Ig , result of different forms of the same gene, involve changes of 1-3 AA in constant region; known allotypes IgG: Gm (Ig Markers) Km (markers on k chain Idiotype = one of several thousand Ig, each of which directed toward specific epitope