* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Weak interactions - Digilander
Expression vector wikipedia , lookup
Signal transduction wikipedia , lookup
Magnesium transporter wikipedia , lookup
Biosynthesis wikipedia , lookup
Gene expression wikipedia , lookup
Genetic code wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Point mutation wikipedia , lookup
Structural alignment wikipedia , lookup
Metalloprotein wikipedia , lookup
Interactome wikipedia , lookup
Biochemistry wikipedia , lookup
Two-hybrid screening wikipedia , lookup
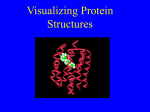
Operone lac Principles of protein structure and function • Function is derived from structure • Structure is derived from amino acid sequence • Different activities and shapes of proteins due to different amino acid sequences Four levels of protein structure Primary: amino acid sequence Four levels of protein structure Primary: amino acid sequence Secondary: regular, repeated coiling and folding of polypeptide backbone Four levels of protein structure Primary: amino acid sequence Secondary: regular, repeated coiling and folding of polypeptide backbone Tertiary: complete three-dimensional structure Quaternary: arrangement of subunits (in multisubunit protein) Secondary structure • Regular, repeated coiling and folding of polypeptide backbone • Due to hydrogen bonding • Two patterns – (alpha) helix – (beta) sheet Tertiary • Complete three-dimensional structure • Due to weak interactions between side (R) groups as well as covalent disulfide bonds Weak interactions Hydrogen bonds Electrostatic interactions (ionic bonds) Hydrophobic interactions Van der Waals interactions Tertiary structure formed through side chain interactions Hydrophobic interactions Tertiary • Complete three-dimensional structure • Composed of: – Motifs: specific combinations of secondary structural elements – Domains: structurally independent units Motifs specific combinations of secondary structural elements Domains Structurally independent units Two different binding domains to bind two different molecules Tertiary • Complete three-dimensional structure Native conformation: functional structure Most stable conformation Tertiary Fibrous Proteins = extended filaments or Globular proteins = compact folded structure Protein Folding • Forming polypeptide chain requires energy and information (template) Protein Folding • Forming polypeptide chain requires energy and information (template) • Forming native conformation requires NO additional energy or information (SELF ASSEMBLY) Protein folding Amino acid sequence contains all information necessary for folding into a specific threedimensional structure Protein Folding Protein Folding Many proteins fold by Assisted Self Assembly Correct assembly (native conformation) requires assistance by CHAPERONES Protein unfolding = Denaturation Loss of structure and function – – – – Heat Extreme pH Detergents Urea Protein unfolding = Denaturation Why do these conditions cause loss of structure and function? – – – – Heat Extreme pH Detergents Urea Tertiary • Complete three-dimensional structure • Due to weak interactions between side (R) groups as well as covalent disulfide bonds Lysozyme Lysozyme Tertiary: complete three-dimensional structure Quaternary: arrangement of subunits (in multisubunit protein) Folding dell’emoglobina Ribbon diagram Hemoglobin Quaternary structure • Held together by weak interactions between side (R/functional) groups as well as covalent disulfide bonds Structure-function relationship • Function is derived from structure • Structure is derived from sequence Sickle-cell disease Normal red blood cells Sickle shaped red blood cells Due to single amino acid change in hemoglobin = protein carries oxygen in red blood cells Sickle-cell disease Sickle-cell disease • Single specific amino acid change causes change in protein structure and solubility • Results in change in cell shape • Causes cells to clog blood vessels Structure-function relationship • Function is derived from structure • Structure is derived from sequence • Molecules with similar shapes can interact in similar ways. – For example, morphine, heroin, and other opiate drugs are similar enough in shape that they can bind to the same receptors as natural signal molecules, called endorphins. – Binding to the receptors produces euphoria and relieves pain. Fig. 2.19 Copyright © 2002 Pearson Education, Inc., publishing as Benjamin Cummings Structure-function relationship • Function is derived from structure • Structure is derived from sequence › Similar sequences have similar functions Structure-function relationship • Function is derived from structure • Structure is derived from sequence › Similar sequences have similar functions › Similar function often implies evolutionary relatedness (homology) » Sequence similarity suggests common evolutionary origin Homologous proteins • Similar sequence • Similar structure • Similar function • Evolved from common ancestor • Belong to protein family Proteins Protein modification and degradation 1. Covalent chemical modification 2. Proteolytic processing and protein splicing 3. Proteolytic degradation Reading and questions • Chapter 3 • Questions 3-1, 3-2, 3-8, 3-11a, 3-15a -15d -15e, 3-16