* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 6. 3-D structure of proteins
Bimolecular fluorescence complementation wikipedia , lookup
Implicit solvation wikipedia , lookup
Protein design wikipedia , lookup
Rosetta@home wikipedia , lookup
Protein purification wikipedia , lookup
List of types of proteins wikipedia , lookup
Western blot wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Structural alignment wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Homology modeling wikipedia , lookup
Protein folding wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein domain wikipedia , lookup
Metalloprotein wikipedia , lookup
Circular dichroism wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
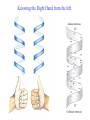
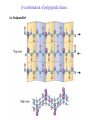
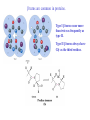
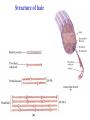
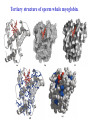
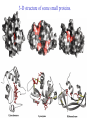
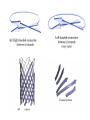
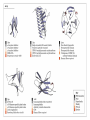
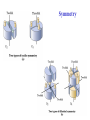
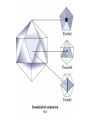
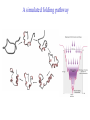
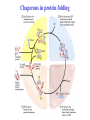
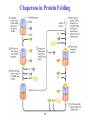
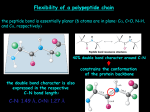
3-D Structure of Proteins • The spatial arrangement of atoms in a protein is called its conformation. • Proteins in any of their functional folded conformations are called native proteins. • Stability can be defined as the tendency to maintain a native confirmation. • When water surrounds a hydrophobic molecule, the optimal arrangement of hydrogen bonds results in a highly structured shell or solvation layer of water in the immediate vicinity. By convention the bond angles resulting from rotations at Cα are labelled Φ (phi) for the N-Cα bond and Ψ (psi) for the Cα-C bond. By convention Φ and Ψ are defined as 180o when polypeptide is in its fully extended conformation and all peptide groups are in the same plane. Ramachandron Plot Allowed values for the Φ and Ψ are graphically revealed when Ψ is plotted versus Φ in a Ramachandran plot. 3-D Structure of Proteins • Primary Structure – basic amino acid sequence • Secondary Structure – refers to the local conformation of some part of the polypeptide [α helix; β conformations] • Teritiary Structure - is the overall three-dimensional arrangement of all atoms in a protein. • Quaternary Structure – is the three-dimensional complexes of protein subunits. α helix – different aspects of its structure α helix – Ψ = -45o to -50o; Φ = -60o Each helical turn includes 3.6 amino acid residues The helical twist of α helix found in all proteins is right-handed. Knowing the Right Hand from the left. Not all polypeptides can form a stable α helix. Interactions between amino acid side chains can stabilize or destabilize this structure. Five different kinds of constraints affect the stability of an α helix: 1. the electrostatic repulsion (or attraction) between successive amino acid residues with charged R group. 2. the bulkiness of adjacent R group 3. the interactions between amino acid side chains spaced three (or four) residues apart. 4. the occurrence of Pre and Gly residues. 5. the interaction between amino acid residues at the ends of the helical segment and the electric dipole inherent of the α helix. β-conformation of polypeptide chains. The structures are somewhat similar, although the repeat period is shorter for the parallel conformation (6.5Å versus 7Å for anti-parallel) and the hydrogenbonding patterns are different. β turns are common in proteins. Type I β turns occur more than twice as frequently as type II. Type II β turns always have Gly as the third residue. Ramachandran plot for a variety of structures. Structure of hair. Structure of Collagen. • Like all the α-keratins, collagen has evolved to provide strength. • Found in connective tissue such as tendons, cartilage, the organic matrix of bone, cornea of the eye. • It is left-handed and had three amino acid residues per term. Structure of Silk Tertiary structure of sperm whale myoglobin. X-ray Diffraction. 3-D structure of some small proteins. • Supersecondary structures, also called motifs or simply folds, are particularly stable arrangements of several elements of secondary structure and the connections between them. • Class and Fold – are purely structural • Family – similar structure and function • Superfamily – little primary sequence similarity, but make use of same major structural motif and have functional similarity α and β segments are interspersed or alternate α and β regions are somewhat segregated Quaternary Structure • Multimer – multi-subunit protein – from 2 to 100 subunits • Oligomer – multimer with few subunits • Protomer – multimer with repeating structural unit Symmetry Renaturation of unfolded, denatured ribonuclease A simulated folding pathway Chaperons in protein folding Chaperons in Protein Folding