* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Genetics in the Generation of Antibody Diversity
Survey
Document related concepts
Transcriptional regulation wikipedia , lookup
Gene desert wikipedia , lookup
Non-coding DNA wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genomic imprinting wikipedia , lookup
List of types of proteins wikipedia , lookup
Gene regulatory network wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Monoclonal antibody wikipedia , lookup
Point mutation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Genome evolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene expression profiling wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Community fingerprinting wikipedia , lookup
Molecular evolution wikipedia , lookup
Transcript
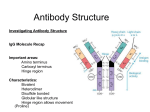
Genetics in the Generation of Antibody Diversity Folder Title: AbGenes(NoTP) Updated: October 2, 2016 Chapter Seven, 7th Edition: “Organization and Expression of Lymphocyte Receptor Genes” Includes Immunoglobulin and T-Cell Receptor Genetics Questions About B-Cell Responses Biochemical Questions: (Slides 3 to 8 Review: Fab Variable Region Sequences) 1. How can Immunoglobulins recognize so many different epitopes? 2. What does an antibody protein look like? 3. How do specific antibodies work as proteins binding so many different specific antigenic determinants? Representation of Sequence Comparisons Among Light Chains from Antibodies with Three Different Antigen Specificities H3N-Ser-Val-Ile-Thr-Gly-Gly-Tyr-Ala... Thr-Glu-Ala-Val-Tyr-Ser-Met-COOH3N-Ser-Ile-Met-Thr-Arg-Leu-Tyr-Gly..Thr-Glu-Ala-Val-Tyr-Ser-Met-COOH3N-Thr-Gly-Gly-Thr-Lys-Leu-Tyr-Ile..Thr-Glu-Ala-Val-Tyr-Ser-Met-COO- Variable Amino Terminal Half Conserved Carboxyl Terminal Half (Positions 1 to 107) (Positions 108 - 214) LiteComp See Figure 4-6, Kuby 6th Edition p. 85 Variable Regions Including hypervariable CDR’s (Complementarity determining regions) κ or λ forms See Figure 4-6, Kuby 6th Edition p. 85 Heavy Chain Iso-forms The Heavy and light chains are labeled incorrectly in the Kuby Immunology Powerpoint slides. The figure is labeled correctly in the book. Questions About B-Cell Responses 1. Biochemical Questions: How can Immunoglobulins recognize so many different epitopes? 2. 3. What does an antibody protein look like? How do specific antibodies work as proteins binding so many different specific antigenic determinants? Genetic Questions: How can this diversity of structure leading to enormous diversity of function be coded and controlled by a very limited host genome? How can we have “one gene - one polypeptide” and make a virtually limitless selection of polypeptides? Problems for Genetics in Generating Antibody Sequence Diversity Vast Sequence Diversity- Encoded by Very Limit Genome Heavy and Light Chain Sequence VariationsAlmost Exclusively in the Amino Terminal One Half of the Light Chains and the Amino Terminal One fourth of the Heavy Chains Exactly the same V-region sequencesEnd up on different C-region Isotypes. How is all of this possible????? Differentiated Neoplastic Plasma Cell making a single antibody. Myeloma Cell DNA Myeloma Cell Conclusions Variable and conserved regions of light chain are linked in the differentiated end-product cell line DNA coding for the mRNA for the light chain is all in one Continuous sequence as for any gene for any protein. Embryonic Mouse Cell DNA Variable and conserved regions of light chain are not linked originally in the stem cell lineage! Immunology and Phone Numbers 315-443-1870 212-345-1775 478-367-8903 537-503-2078 409-159-6309 610-970-3970 934-620-8122 909-603-7023 800-620-6021 704-590-5307 703-725-0153 207-502-6671 435-431-0890 412-830-0048 740-592-1954 307-620-4450 490-501-5672 601-909-7002 554-891-7712 335-592-0944 20 Phone Numbers Immunology and Phone Numbers 315212478537409610934909800704703207435412740307490601554335- -443-1870 -345-1775 -367-8903 -503-2078 -159-6309 -970-3970 -620-8122 -603-7023 -620-6021 -590-5307 -725-0153 -502-6671 -431-0890 -830-0048 -592-1954 -620-4450 -501-5672 -909-7002 -891-7712 -592-0944 400 Phone Numbers Immunology and Phone Numbers 315212478537409610934909800704703207435412740307490601554335- -443-345-367-503-159-970-620-603-620-590-725-502-431-830-592-620-501-909-891-592- -1870 -1775 -8903 -2078 -6309 -3970 -8122 -7023 -6021 -5307 -0153 -6671 -0890 -0048 -1954 -4450 -5672 -7002 -7712 -0944 8000 Phone Numbers 41 Kappa V-Region Sequences in Humans “Pseudo-gene” is not expressed Kappa Chain DNA: Vk’s This and the next two slides deal with Kappa Light Chain DNA. Human Kappa DNA was 41 different “V” region genes to work with, and 4 “J” region genes. (164 Possibilities) Lambda light chain DNA works the same way except that the Human Lambda light chain works with 33 “V” regions and 5 “J” region genes. (165 Possibilities) Kappa Chain: K-J- C Complete Kappa Chain Light chain V-region genes do not have diversification genes Heavy Chain Dna: VH’s, CGenes This and the next two slides deal with Heavy chain DNA. Human heavy chain DNA has 41 “V” region genes to work with, 23 “D” (diversification) region genes, and 6 “J” (joining) region genes. Heavy chain V and J region genes are different from Light Chain V and J region genes and are on different chromosomes from the Light Chain genes. (Chromosome 14 in Humans for Heavy Chain genes. Chromosomes 2 and 22 for Light Chain genes (Kappa and Lambda) 5658 Possibilities even with no additional variations Coupling a VH option with DH option and a JH option to a Cu gene sequence would give a Mu Heavy chain isotype with a specific antibody recognizing specificity. Coupling that exact same VH-DH-JH genetic information to Cd or Cg or Ce or Ca Heavy chain isotype gene would give the exact same antigen specificity on a different isotype. We would have Isotype Switching Chain Structures of the five immunoglobulin classes in humans (adapted from Kuby, 2nd edition) Class Heavy Light Sub-Classes Subunit Formula Chain Chain IgA γ α κ or λ γ1, γ2, γ3, γ4 γ2κ2 γ2λ2 κ or λ α1, α2 (α2κ2)n (α2λ2)n n =1,2,3,4 IgM μ κ or λ None IgD δ κ or λ None δ2κ2 δ2λ2 IgE ε κ or λ None ε2κ2 ε2λ2 IgG (μ2κ2)n (μ2λ2)n n = 1 or 5 Complete Heavy Chain The puzzle of immunoglobulin gene structure Kappa or Lambda Light Chains • B cells use groups of PARTS of genes to create different possible antibodies using recombination – Like shuffling a deck of cards, dealing out different hands – Tightly regulated machinery controls the recombination processes The puzzle of immunoglobulin gene structure • B cells use groups of PARTS of genes to create different possible antibodies using recombination – There are variable (V), diversity (D), joining (J), and constant (C) region gene segments • D segments are used in antibody heavy chains only Multigene organization of Ig genes • Recall that Ig proteins consist of: – two identical heavy chains – two identical light chains • Light chains can be either kappa or lambda – Each set of gene families are encoded on separate chromosomes The mechanism of V(D)J recombination • Five mechanisms generate antibody diversity in naïve B cells – Multiple gene segments―which gene segments are put together – P nucleotide addition―templated nucleotide addition between joints, resulting from assymetrical cleaving of hairpin structures – Exonuclease trimming―sometimes occurs at junctions, losing nucleotides and changing reading frames – Non-templated N nucleotide addition―mediated by TdT activity, adding in random nucleotides between joints - Somatic Hypermutation Combinatorial diversity―which heavy chains pair with which light chains The mechanism of V(D)J recombination • Five mechanisms generate antibody diversity in naïve B cells (See previous slide) One pseudo gene Additional sequence diversity estimated at several orders of magnitude (i.e. perhaps 1000-fold) is generated by junctional flexibility, nucleotide addition, and somatic hypermutation. This allows for an estimated 100 million to a billion different possible antibody seuquences in humans. B-Cell Differenatiation B-cell receptor expression • Allelic exclusion ensures that each B cell synthesizes only one heavy and one light chain. Recombination is a very ordered process. Nonproductive arrangements lead to programmed cell death (apoptosis) during development. Once one successful heavy chain allele has been assembled, the other allele is shut off. Cell then goes to making a light chain. Once one successful kappa or lambda light chain has been assembled, the other kappa or lambda allele is shut off. Detailed genetic mechanisms for generating antibody diversity are shown in Chapter 7 of Edition 7. These specifics are beyond the scope of this introductory course but are important for those interested in advanced work in immunobiology or medical genetics. Please put away all notes and devices other than your XR transmitter. No consulting or reading notes is permitted. No online searches. This are graded response questions. Rank Responses 17% 17% 17% 17% 17% 17% 1 2 3 4 5 6 Response Counter 1 2 3 4 5 6 ___ ___ ___ ___ ___ ___ ____. switching. Rank Responses 1 2 3 4 5 Response 6 Counter 0% 0% 1 2 0% 0% 3 4 0% 0% 5 6 17% 17% 17% 5 Rank 1 2 17% 4 17% 17% Responses 3 4 6 3 2 Response Counter 1 5 6 Save Session with Participant List Responses. Close Session. Reset to Anonymous Polling Open New Session for Anonymous Slide on Scope and Pace of the course so far. This question will be set to anonymous so no one will know what any specific student says. For the first 11 classes the pace of the course is: 1. Too slow. We need to move faster and cover more about Immunology. 2. About right. I can keep up Ok but it is a challenge. 3. Way too fast. Slow down! We’ll figure out later what you don’t get to cover in class. 0% Response Counter 1. 0% 2. 0% 3. On a scale of 5 to 1 rate todays AbGenes Presentation 5. Too slow. Get moving 4. No problem. 3. About right. I can figure out what I missed. 2. Too fast 1. I’m lost! (This is anonymous. We won’t know who you are.) 5 4 3 2 1 Duration: 0 Seconds