* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Slides
Endogenous retrovirus wikipedia , lookup
Gene regulatory network wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Silencer (genetics) wikipedia , lookup
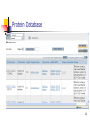
Gene expression wikipedia , lookup
Expression vector wikipedia , lookup
Peptide synthesis wikipedia , lookup
Gene nomenclature wikipedia , lookup
Interactome wikipedia , lookup
Magnesium transporter wikipedia , lookup
Metalloprotein wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein purification wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Genetic code wikipedia , lookup
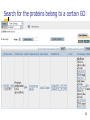
Biosynthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Proteins and Protein Function Charles Yan Spring 2006 Amino Acids General structure of an amino acid 20 standard amino acids each with a different R group 2 Amino Acids Table 1. 20 standard amino acids Amino Acid 3-letter code 1-letter code Alanine Ala A Arginine Arg R Asparagine Asn N Aspartate Asp D Cysteine Cys C Glutamine Gln Q Glutamate Glu E Glycine Gly G Histidine His H Isoleucine Ile I 3 Amino Acids Table 1. 20 standard amino acids (Cont.) Amino Acid 3-letter code 1-letter code Leucine Leu L Lysine Lys K Methionine Met M Phenylalanine Phe F Proline Pro P Serine Ser S Threonine Thr T Tryptophan Trp W Tyrosine Tyr Y Valine Val V 4 Amino Acids Amino Acid 3-letter code 1-letter code Asparagine (N) or aspartate (D) Asx B Glutamine (Q) or glutamate (E) Glx Z Any amino acid Xaa X Amino Acid Abbreviations (IUPAC) Authority Reference IUPAC-IUB Joint Commission on Biochemical Nomenclature. IUPAC-IUB Joint Commission on Biochemical Nomenclature. Nomenclature and Symbolism for Amino Acids and Peptides. Eur. J. Biochem. 138:9-37(1984). 5 Proteins Two separate amino acids can be linked together by a peptide bond A chain of amino acids linked by peptide bonds is called a polypeptide. A protein is made up of one or more polypeptide chains For simplicity, in this course, a protein is a chain of amino acids linked by peptide bonds, e.g. VSQLLKQRVRYAPYLSKVRRAEELLPLFKHGQYIGWSGFTGVGAPKVI 6 Protein Database UniProt (Universal Protein Resource) (http://www.pir.uniprot.org/) is the world's most comprehensive catalog of information on proteins. It is a collaboration between Swiss Institute of Bioinformatics (SIB) Department of Bioinformatics and Structural Biology of the Geneva University European Bioinformatics Institute (EBI) Georgetown University Medical Center's Protein Information Resource (PIR) It includes three components 7 Protein Database UniProt Knowledgebase (UniProtKB): the central access point for extensive curated protein information. UniProtKB/Swiss-Prot: a manually annotated protein sequence database which provide a high level of annotation, a minimal level of redundancy and high level of integration with other databases. UniProtKB/Swiss-Prot Release 48.7 of 20-Dec-2005: 204,086 entries UniProtKB/TrEMBL: a computer-annotated supplement of Swiss-Prot that contains all the translations of EMBL nucleotide sequence entries not yet integrated in Swiss-Prot. UniProtKB/TrEMBL Release 31.7 of 20-Dec-2005: 2,506,886 entries UniProt Reference Clusters (UniRef): databases combine closely related sequences into a single record to speed searches. UniProt Archive (UniParc): a comprehensive repository, reflecting the history of all protein sequences 8 Protein Database 9 Protein Database 10 Protein Database 11 Protein Database 12 Protein Database 13 14 Gene Ontology Goal: find all the proteins that are involved protein synthesis Protein synthesis Translation 15 Gene Ontology Volkswagen Golf I like golf. Me too! Golf 16 Gene Ontology Ontology n. the branch of metaphysics dealing with the nature of being. (The New Oxford American Dictionary, Edited by Elizabeth J. Jewell, Frank Abate, Oxford University Press, 2001,pp 1197.) Metaphysics n. the branch of philosophy that deals with the first principles of things, including abstract concepts such as being, knowing, substance, cause, identity, time, and space. (The New Oxford American Dictionary, Edited by Elizabeth J. Jewell, Frank Abate, Oxford University Press, 2001,pp 1074.) 17 Gene Ontology The Gene Ontology (GO) (http://www.geneontology.org/) project is a collaborative effort to address the need for consistent descriptions of gene products in different databases. The project began as a collaboration between three model organism databases: FlyBase (Drosophila),the Saccharomyces Genome Database (SGD) and the Mouse Genome Database (MGD) in 1998. Since then, the GO Consortium has grown to include many databases, including several of the world's major repositories for plant, animal and microbial genomes. 18 Gene Ontology Develop structured, controlled vocabularies (ontologies) that describe gene products Make associations between the ontologies and the genes and gene products in the collaborating databases, Develop tools that facilitate the creation, maintainence and use of ontologies The use of GO terms facilitates uniform queries across databases 19 Gene Ontology The three components of GO are molecular function, biological process and cellular component GO terms are organized in structures called directed acyclic graphs (DAGs), which differ from hierarchies in that a child, or more specialized, term can have many parent, or less specialized, terms monosaccharide biosynthesis hexose metabolism hexose biosynthesis 20 Gene Ontology The controlled vocabularies are structured so that you can query them at different levels GO browser AmiGO (http://www.godatabase.org/cgibin/amigo/go.cgi) 21 22 Protein function Three steps to get a set of proteins that have a certain function Search for the GO term (http://www.godatabase.org/cgi-bin/amigo/go.cgi) Search for the proteins belong to a certain GO (http://www.pir.uniprot.org/search/textSearch.shtml) Save the sequence in FASTA format 23 Search for the GO 24 Search for the proteins belong to a certain GO 25 Save sequences in FASTA format 26