* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download CMSE 520 BIOMOLECULAR STRUCTURE, FUNCTION AND
Non-coding DNA wikipedia , lookup
Metalloprotein wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Drug design wikipedia , lookup
Signal transduction wikipedia , lookup
Molecular ecology wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Size-exclusion chromatography wikipedia , lookup
Expression vector wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression wikipedia , lookup
Western blot wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Proteolysis wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Interactome wikipedia , lookup
Two-hybrid screening wikipedia , lookup
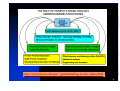
CMSE 520 BIOMOLECULAR STRUCTURE, FUNCTION AND DYNAMICS (Computational Structural Biology) OUTLINE Review: Molecular biology Proteins: structure, conformation and function(5 lectures) Generalized coordinates, Phi, psi angles, DNA/RNA: structure and function (3 lectures) Structural and functional databases (PDB, SCOP, CATH, Functional domain database, gene ontology) Use scripting languages (e.g. python) to cross refernce between these databases: starting from sequence to find the function Relationship between sequence, structure and function Molecular Modeling, homology modeling Conservation, CONSURF Relationship between function and dynamics Confromational changes in proteins (structural changes due to ligation, hinge motions, allosteric changes in proteins and consecutive function change) Molecular Dynamics Monte Carlo Protein-protein interaction: recognition, structural matching, docking PPI databases: DIP, BIND, MINT, etc... References: CURRENT PROTOCOLS IN BIOINFORMATICS (e-book) (http://www.mrw.interscience.wiley.com/cp/cpbi/articles/bi0101/frame.html) Andreas D. Baxevanis, Daniel B. Davison, Roderic D.M. Page, Gregory A. Petsko, Lincoln D. Stein, and Gary D. Stormo (eds.) 2003 John Wiley & Sons, Inc. INTRODUCTION TO PROTEIN STRUCTURE Branden C & Tooze, 2nd ed. 1999, Garland Publishing COMPUTER SIMULATION OF BIOMOLECULAR SYSTEMS Van Gusteren, Weiner, Wilkinson Internet sources Ref: Department of Energy Rapid growth in experimental technologies Human Genome Projects Two major goals 1. DNA mapping 2. DNA sequencing Rapid growth in experimental technologies z Microrarray technologies – serial gene expression patterns and mutations z Time-resolved optical, rapid mixing techniques - folding & function mechanisms (Æ z Techniques for probing single molecule mechanics (AFM, STM) (Æ pN) Æ more accurate models/data for computer-aided studies Weiss, S. (1999). Fluorescence spectroscopy of single molecules. Science 283, 1676-1683. ns) function Structural Biology/Molecular Biophysics Most (all?) basic “life processes” are mediated by “machines” that represent the ultimate miniaturization achievable in a universe comprised of atoms and molecules. The goal is to understand the underlying principles that govern the operation of these molecular machines. What th is course is about this overview of ways in which computers are used to solve problems in biology supervised learning of illustrative or frequently-used algorithms and programs and databases supervised learning of programming techniques and algorithms selected from these uses Structure What do the molecules look like? How do we determine that experimentally? Are there general structural principles? How is this information organized? How do structural generalizations relate to simple physical/chemical principles? Dynamics Time is of the essence in biological processes therefore how do we understand time-dependent processes at the molecular level? How do we do this experimentally? How do we do this computationally? Promising Future for Computational Biology Exponential growth in data Sequence and structure data from experiments Computational technology 12,665 structures as of July 11, 2000 22,810 structures as of October 7, 2003 35,026 structures as of February 7, 2006 Rost, B. (1998). Marrying structure and genomics. Structure 6, 259-263 Large databases Archival databanks of biological information Protein, DNA sequence databases Protein structure and nucleic acid databases Protein expression patterns Experimental Tecniques Derived databanks Sequence motifs Mutations and variations in proteins Classifications and or relationships Databanks of web sites Databanks of databanks containing biological information Links between databanks BIOINFORMATICS (definition) Definition by Luscombe et al., Yale, Dept. of Molecular Biophysics and Biochemistry, 2001 “Bioinformatics is conceptualizing biology in terms of macromolecules (in the sense of physical chemistry) and then applying ‘informatics’ techniques (derived from disciplines such as applied maths, computer science, and statistics) to understand and organize the information associated with these molecules, on a large-scale” COMPUTATIONAL BIOLOGY (definition) Definition by NIH (working definition) The development and application of data-analytical and theoretical methods, mathematical modeling and computational simulation techniques to the study of biological, behavioral, and social systems. Information flow A major task in computational molecular biology is to “decipher” information contained in biological sequences Since the nucleotide sequence of a genome contains all information necessary to produce a functional organism, we should in theory be able to duplicate this decoding using computers http://www-fp.mcs.anl.gov/~gaasterland/sg-review-slides.html 5 Two major challenges after completion of the HGP: Structural Genomics and Functional Genomics Schematic representation of the universe of proteins in a given organism Kim, S.H. (1998). Nature Struct.Biol. 5, 643-645 Aim: “to construct the complete scheme of biological functions and cellular pathways for the entire organism” What's E-Cell Project? E-Cell Project is an international research project aiming to model and reconstruct biological phenomena in silico, and developing necessary theoretical supports, technologies and software platforms to allow precise whole cell simulation. Metabolism model of the model cell constructed with 127 genes PROTEOMICS Covers the following areas (but not limited to): ¾Protein structure Primary Structure: sequence of amino acids Secondary Structure: local spatial arrangement Tertiary Structure: three dimensional native conformation ¾Protein Function related to 3-D shape of the protein ¾Protein clusters according to a specified characteristic ¾Protein-Protein Interaction interaction among a number of proteins ¾Protein-DNA Interaction interaction between one protein and the genome